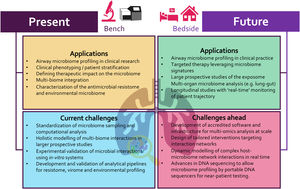
Accelerated by developments in DNA sequencing technologies, our understanding of the respiratory microbiome is advancing at pace, providing unprecedented opportunities for clinical translation.1 Building on the early observations of sub-clinical micro-aspiration in healthy individuals, and initial culture-independent microbiome studies in respiratory disease, recent work reveals an expansive microbial ecosystem that encompasses bacterial, fungal and viral constituents.1–3 This has led to major paradigm shifts including the potential importance of airway microbial networks in chronic respiratory disease states.4 As a complex organ system, with varying topology and a mucosal surface area exceeding that of the gut, the respiratory tract is recognized as a key site of host-microbe interaction. The airway experiences dynamic and continuous microbial exposures on breathing, shaped by climate and environmental surroundings, and is further influenced by sub clinical micro-aspiration of resident upper-airway microbes.5–7 The respiratory microbiome exists as an ecological gradient from upper to lower airway, interacting with host epithelia in balance between immune homeostasis and pathology.8–11 Current models posit that a balanced host-microbe interaction establishes in early life with a protective immune response that become dysregulated in respiratory disease. Characterization of microbial aberration as early indicators of deteriorating respiratory health is therefore a fundamental concept underpinning its potential clinical applications. Detecting microbial dysbiosis from otherwise ‘healthy microbiomes’ represents a potential opportunity for personalized phenotyping, stratification and therapeutic intervention (Fig. 1). Despite such promise, this relatively nascent field has inherent challenges that need addressing as we seek to translate research gains in our understanding of the airway microbiome into tangible clinical applications for respiratory medicine (Fig. 1).
The airway microbiome: present applicationsIn contrast to pathogen-centric, culture-based studies, we are now equipped to identify complex microbial networks influencing lung inflammation and tissue injury at high resolution. Integrative methods have allowed incorporation of bacteriomes, mycobiomes and viromes in the generation of individual patient ‘multi-biome’ profiles.1,4,12 The respiratory microbiome is now well described in several chronic respiratory disorders including asthma, chronic obstructive pulmonary disease (COPD), bronchiectasis and cystic fibrosis (CF), as well as in the infectious disease, critical care and transplant settings.1,4,13,14 These studies, despite being largely observational, highlight the ability to stratify patients based on microbiome profiles with strong association to clinical phenotype. Assessing microbiome responses to therapy, including antibiotics, has surprisingly illustrated minimal profile change to individual microbes, while microbial interaction networks appear more promising predictors of a therapeutic response.4 Work to date on phenotyping respiratory disease has predominantly employed 16S ribosomal RNA sequencing which characterizes bacteriomes but excludes other microbial communities including the mycobiome and virome respectively, leading to bias. To address this issue, our group has undertaken targeted mycobiome analyses in bronchiectasis and COPD revealing an alternate perspective. Fungi have an important role to play in defining patient outcome: they can identify high-risk COPD and associate with sensitization and Allergic Bronchopulmonary Aspergillosis (ABPA) in bronchiectasis.10,14 Furthermore, integration of mycobiomes with corresponding bacteriome and virome profiles improves precision for identifying ‘at-risk’ patient groups underscoring the importance of inter-kingdom networks in microbiome medicine.4 Other key insights provided by microbiome analyses (through whole genome shotgun metagenomics) includes functional (pathway) assessment, appraisal of the antimicrobial resistome and capturing environmental exposures, some of which have been correlated to clinical outcomes in respiratory disease.7,15
A key goal of microbiome research is its incorporation into diagnostics or as a point-of-care tool for clinical benefit. While technically feasible, significant challenges remain, notably in sampling the heterogenous ecology of the respiratory tract, which itself is influenced by changing micro-environments such as pH, surfactant production, muco-ciliary clearance, and the availability of nutrients. The presence of chronic respiratory disease coupled to its geographically variable phenotypes adds further complexity. Standardization of microbiome sequencing while somewhat established for 16S rRNA gene analysis, is less adapted for fungal ITS analysis, while airway virome sequencing approaches remain in their infancy. There has been limited advance in understanding composition of the respiratory virome (including bacteriophages) and its potential interaction with other airway constituents at the functional and metabolic level. Standardization and validation of computational approaches for analysis represents another key hurdle to the widespread clinical implementation of airway microbiome assessment. Access to analytical pipelines capable of handling complex data in an efficient, cost-effective, and reproducible manner is critical to execution at scale in a clinical or near-patient setting.
The airway microbiome: future applicationsAs we navigate the airway microbiome at an increasing scale, network-based approaches leveraging multidimensional, integrative, and dynamic characterization will deepen our current understanding. These approaches better approximate ‘real-world’ states characterizing the respiratory microbiome and its constituents. Once considered an ambitious concept attainable only by specialist groups, an integrative assessment incorporating relevant multi-biomes (i.e. bacterial, fungal, and viral) including assessment of varying sites (e.g. lung and gut) now represents an achievable endpoint with potentially amenable results for clinical application. Integration of lung and gut profiles is of particular interest given the emerging concept of a ‘lung-gut’ axis, generating additional diagnostic precision that exceeds a single-organ ‘lung-only’ microbiome view.1,4 The application of such microbiome assessment in prospective studies (including clinical trials) is critical to better our understanding of optimal patient stratification, identifying ‘responders’ early and ensuring a consideration of disease-specific microbial complexity. In future, we even need to move beyond such multi-biome and multi-site microbiome profiling and include an external analysis of the ‘exposome’ that incorporates the air microbiome in residential and work environments.7 Finally, the use of multi-omics technologies and high-throughput profiling of the host immune response can further stratify patients and their clinical phenotypes to specific microbial exposures and offer avenues for therapeutic intervention.11 The integration of host ‘omics’ profiles to microbiome analysis will provide even deeper insight to endo-phenotyping efforts and allow potential prediction of patient trajectory and/or treatment response in the emerging era of precision medicine.
Given the ‘data-rich’ nature of airway microbiome research and the emergence of new platforms and technologies, it is critical that computational and analytic development keeps pace. Software solutions must be reproducible, efficient, and user-friendly to prevent over-reliance on specialist expertise, a key hurdle to clinical translation. Novel sequencing platforms may help to reduce such technical burdens and ‘democratize’ technologies to ease translation. While the capacity of DNA sequencing platforms continues to excel, novel technologies such as portable nanopore sequencing represent a potential avenue for near patient assessment of airway microbiomes. While bacteriomes are well characterized in several respiratory studies, significantly more work on the mycobiome and virome (including bacteriophages) is needed and metagenomic approaches should be leveraged. Integrating host immunology and genetics with airway microbiome profiles remains a challenge, one that requires data integration that accurately incorporates a multi-organ capture of multi-biomes with carefully designed discovery and validation cohorts.
As a respiratory community, our significant progress in airway microbiome research over the last decade has provided opportunities for translation to clinical care. To realize this over the next decade will require addressing its challenges including leveraging the innovation in rapidly progressing analytical techniques while translating such data-rich capture into clinical insight in a cost-effective, scalable, applied, and reproducible manner. Precision ‘microbiome’ medicine is the ultimate aspiration, one that now lies at the frontier of airway microbiome research and clinical care.