Chronic obstructive pulmonary disease (COPD) is an entity with a heterogeneous presentation. For this reason, attempts have been made to characterize different phenotypes and endotypes to enable a more individualized approach. The aim of the Biomarkers in COPD (BIOMEPOC) project is to identify useful biomarkers in blood to improve the characterization of patients. Clinical data and blood samples from a group of patients and healthy controls will be analyzed. The project will consist of an exploration phase and a validation phase. Analytical parameters in blood will be determined using standard techniques and certain ‘omics’ (transcriptomics, proteomics, and metabolomics). The former will be hypothesis-driven, whereas the latter will be exploratory. Finally, a multilevel analysis will be conducted. Currently, 269 patients and 83 controls have been recruited, and sample processing is beginning. Our hope is to use the results to identify new biomarkers that, alone or combined, will allow a better characterization of patients.
La enfermedad pulmonar obstructiva crónica (EPOC) es una entidad de presentación heterogénea. Por ello, se han intentado perfilar diferentes fenotipos y endotipos, que permitirían un manejo más diferenciado. El objetivo del proyecto Biomarcadores en la EPOC (BIOMEPOC) es identificar biomarcadores sanguíneos útiles para tipificar mejor a los enfermos. Se analizarán datos clínicos y muestras sanguíneas en un grupo de pacientes y controles sanos. El proyecto constará de fases de prospección y de validación. Se realizarán determinaciones analíticas sanguíneas con técnicas convencionales y de diversas ciencias «ómicas» (transcriptómica, proteómica y metabolómica). Las primeras se realizarán orientadas por hipótesis, mientras que con las segundas se realizará una exploración sin dicho condicionante. Finalmente se realizará un análisis multinivel. En el momento actual se han reclutado 269 pacientes y 83 controles, y se está iniciando el procesamiento de muestras. Con los resultados obtenidos se espera identificar nuevos biomarcadores que, en solitario o combinados, permitan una mejor tipificación de los pacientes.
Chronic obstructive pulmonary disease (COPD) is a clinical entity characterized by a decline in lung function that is frequently progressive with intermittent exacerbations of symptoms.1,2 Prevalence is very high, and the disease generates considerable social and healthcare costs.2,3 The main cause of COPD is smoking, although other factors may contribute to its appearance.2,4 An important characteristic of this entity is that its presentation is very heterogeneous: pulmonary involvement typically varies widely, and is often accompanied by extrapulmonary manifestations and various comorbidities.5,6 The most notable extrapulmonary effects include compromise of the patient's nutritional status and musculoskeletal system, while significant comorbidities include cardiovascular and endocrine disorders and lung cancer. These comorbidities are often compounded by the negative effects of aging (since exposure to the toxic substance is usually prolonged before clinical symptoms of the disease manifest), and in many cases by an unfavorable social environment (which affects the more vulnerable segments of society in particular). Alongside this understanding of COPD as a complex and heterogeneous entity, FEV1 has been established as the variable that reflects the intensity of airflow obstruction. Until recently, FEV1 has been the only parameter used for classifying severity among patients, but it provides little information on prognosis and appropriate clinical management and treatment, since other elements that are highly significant in terms of the disease profile are not taken into account.
These considerations have led in recent years to a search for more specific patient profiles, in an attempt to offer a more individualized approach to clinical care, treatment, and continued follow-up. In this respect, the GOLD classification recently incorporated an evaluation of symptoms and the annual number of exacerbations, along with the now standard FEV1.2 In a further step, other groups have attempted to define more specific profiles, proposing the term “phenotype”, used rather differently to the traditional meaning, as it refers here to the clinical or biological features that might affect the prognosis or therapeutic management of patients.7 In this respect, new classifications have been added to the 2 conventional disease phenotypes (pulmonary emphysema and chronic bronchitis), and phenotypes such as “frequent exacerbator” and “mixed” (e.g., asthma-COPD overlap [ACO]) have been included.8,9 Other groups have described various phenotypes based on clinically significant features (e.g., weight loss, cardiovascular comorbidity, or persistent systemic inflammation).10–13 Attempts have also been made to characterize patients based on the pathophysiological mechanisms that explain heterogeneity (endotypes), although this approach is still in its infancy. More recently, it has been proposed that we go even further, with the individualized characterization of patients.13 To apply such personalized medicine, we need to have sufficient indicators to profile each patient as accurately as possible. Not only will the severity of the lung disease have to be defined (based until now on functional residual capacity), but other dimensions, such as degree of activity (intensity of the biological mechanisms that would characterize the endotypes), the impact of the disease on the patient's life,14 and both comorbidities and systemic manifestations will have to be characterized.
On a practical level, this conceptual shift means that new variables (markers) must be identified that, ideally, are easy to obtain and allow patients to be accurately classified. The search for these variables should take into account the heterogeneous and complex characteristics of the entity, and be based on the interrelationship of the various levels that determine disease expression. This, in other words, is a question of addressing the complexity of the disease, in order to achieve an appropriate stratification associated with certain biological characteristics.15 Until now, classification has been based primarily on environmental and clinical factors, but it is time for these to be reinforced with different biological parameters in order to determine the so-called endotypes. This will be only possible by identifying biomarkers, defined as objective indicators of a biological status,15 usually detectable in any of the biological fluids or tissues.16
Unfortunately, attempts so far to determine good biological markers in COPD have not produced the desired results. On the one hand, a great many studies have explored small numbers of molecules, oriented in general toward a hypothesis based on current pathophysiological knowledge (inflammation, oxidative stress, injury or tissue remodeling).17–19 These tend to be studies with relatively large numbers of patients that focus on a specific clinical circumstance (e.g., frequency of exacerbations, rate of progress), and in many cases lack a subsequent validation phase. Results have been inconclusive in general, although the ECLIPSE study led to the observation that the combination of various serum inflammatory markers can help predict progress of the standard clinical variables.17 In recent years, several studies have also been conducted with similar aims, using the so-called “omics” sciences. These techniques are generally characterized by a broad search performed with no specific preset hypothesis. However, these studies have often comprised relatively small series, poorly defined from a clinical point of view, and have therefore also failed to produce the desired results.20–25 The aim of this project is to overcome some of these limitations.
The general hypothesis of the Biomarkers in COPD (BIOMEPOC) project is that the search for biomarkers for the different clinical characteristics typical of COPD (intercurrent exacerbations, degree of activity, presence of eosinophilia, comorbidities, systemic manifestations, etc.) will be much more efficient if standard methodologies and systems biology methodologies are used in combination. This approach will help define interaction networks, by exploring the metabolic pathways and possible molecular mechanisms in each case, ultimately identifying the most appropriate biomarkers. Logically, the specific hypothesis of the “omics” approach will derive from the initial exploration and the molecules identified during this investigation, which will subsequently be validated. In contrast, the specific hypothesis of the part of the study that uses conventional techniques is that the inflammatory markers of oxidative stress and injury-repair in the lung, muscle, and cardiovascular tissues will help distinguish the different forms, phenotypes, and endotypes associated with the pulmonary and systemic manifestations of the disease. Consequently, the general objective of the project is to conduct an integrated analysis of the indicators obtained from the clinical symptoms, standard laboratory tests, and copious multilevel biological data of COPD patients, with the aim of defining new specific profiles and characterizing more accurately the phenotypes and endotypes of those already proposed.
The BIOMEPOC project, with its initial cross-sectional design, focuses on the first 2 of the 4 types of biomarker defined by the European Commission (susceptibility/risk, diagnostic, prognostic, and predictive of treatment response).15 However, future objectives will also include an exploration of the third and fourth types, although this implies a longer follow-up that will go beyond the initial duration of the study.
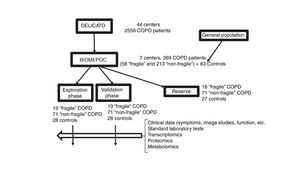
MethodDesign, Population, and Clinical VariablesThe BIOMEPOC study has a prospective design that includes the acquisition of numerous variables and biological samples at the time of recruitment, and the follow-up of some variables such as the subsequent development of lung cancer and mortality. Patients, mostly from a previous study, and healthy controls will be enrolled in the 7 participating centers (Hospital Son Espases, Palma; Fundación Jiménez Díaz, Madrid; Hospital Clínic, Barcelona; Hospital Universitario Virgen del Rocío, Seville; Hospital 12 de Octubre; Madrid; Hospital del Mar, Barcelona, and Hospital Universitari Parc Taulí, Sabadell). Third-level and university hospitals that are accustomed to managing complex patients and processing biological samples that require specialized handling have been selected. The project consists of two phases: an exploratory phase of identifying and selecting biomarkers in a subgroup of patients and healthy controls, and a second phase of internal validation of the results obtained in another subgroup of patients and healthy controls (Fig. 1).
This project would not be possible without the participation of a multicenter group with wide-reaching, long-term experience in collaborative network-based studies, and the availability of a cohort of clinically well characterized patients. Fortunately, we were granted access to a clinical audit study called the “Design and local implementation of Clinical Audits in different types of Obstructive Lung Disease” (DELICATO study), sponsored jointly by the Respiratory Area of the Research Network Center (CIBERES) and the Spanish Society of Pulmonology and Thoracic Surgery (SEPAR), that includes an appropriate cohort for the objectives of this project.
The inclusion and exclusion criteria, then, were the same as those of the DELICATO study. Briefly, patients with COPD2 and a group of healthy controls were studied with continuous follow-up in a specialized respiratory medicine clinic for at least 2 years. We used a standardized questionnaire to recruit healthy controls, well-matched in sex and age with study patients, and with respiratory function tests within the normal range. All subjects were over 40 years of age. Diagnostic criteria for COPD were a history of smoking, and a post-bronchodilator spirometric ratio of less than 70%.2 The sample size was calculated from previous studies,18,26–28 using the study with the largest series26 for the calculation of the total number of COPD patients, and increasing the resulting number generously in anticipation of losses or recruitment difficulties. The other studies were used for the calculation of blood samples to be evaluated in a subanalysis, using the techniques mentioned above. This meant that each of the 7 centers initially had to attempt to recruit 90 COPD patients (about 30 patients considered “fragile” according to the definition adopted in DELICATO: 3 or more hospital visits due to exacerbations in the previous year; and 60 more patients not meeting that criterion) and 30 healthy subjects. All subjects had to provide a blood sample for the biomarker study, and all patients who were able would provide spontaneous sputum sample for further studies.
EthicsThe study was designed on the basis of the principles of the Declaration of Helsinki, and was approved by the Clinical Research Ethics Committees (CREC) of the different institutions. All study participants signed the relevant informed consent form after having been informed in detail of the objectives and procedures to be used. To ensure anonymity, each participant and their samples were allocated a sequential 4-figure number. The general coordinator of the project keeps the link between these numbers and the identity of the subjects in a separate, secure file.
Clinical DataThe clinical, analytical, functional, and follow-up data obtained in the DELICATO project were collected and uploaded to a new centralized database. The presence and severity of emphysema was evaluated from a standard chest X-ray, diffusing capacity of the lung for carbon monoxide was measured, and nutritional status was evaluated from the body mass index. Data from the healthy controls were also added to the database.
Biomarker AnalysisSeveral complementary strategies were used to obtain biomarkers. Firstly, for standard techniques in which the molecules to be analyzed had to be previously defined, parameters were selected from the specialized literature and from a study already published by the study team, using text mining techniques.29 In contrast, as mentioned above, no prior selection was made for comprehensive screening using “omic” techniques, to avoid the potential bias of a predefined hypotheses.
Processing and Analysis of Blood SamplesBefore the project began, the standardized protocol for the collection and initial processing of samples was drafted and distributed among the participants (Appendix B, addendum 1). Samples were sent to the coordinating center and stored in the biobank at −80°C (MARBiobanc). This facility meets the standard criteria for the storage and custody of biological samples (AENOR ER-0031/2012; UNE-EN ISO 90019). The samples will be processed using conventional analytical techniques and comprehensive analysis of transcriptomic, proteomic and metabolomic levels. Part of each sample is reserved for possible future analysis.
- “Conventional” techniques. These include a complete blood count and standard serum determinations and determinations of generic inflammatory markers (ultrasensitive C-reactive protein [CRP], fibrinogen, and glycomarkers), oxidative stress (8-isoprostane, malondialdehyde, reduced glutathione, club-cell [CC] secretory protein-16, and granzime [Gzm] B), infection (procalcitonin), and cardiovascular dysfunction (natriuretic peptide [BNP] and pro-BNP).
- Transcriptome analysis. Samples treated specifically to preserve the ribonucleic acid (RNA) are used. These samples are prepared for massive sequencing (NextSeq, Illumina, San Diego, USA), according to the procedure already described in the literature,30 which can process over 25000 genes per sample. This analysis is performed on the transcriptomics platform of Universidad Pompeu Fabra (UPF) in Barcelona. The results are analyzed by bioinformaticians and clinicians from our group using specific conventional programs, with subsequent functional analysis using the Gene Ontology (GO, Gene Ontology Consortium)31 and Human Phenotype Ontology (HPO, Charité, Berlin, Germany)32 systems. The transcriptome results selected from those which demonstrate a high discriminatory value will be confirmed by the quantitative reverse transcriptase polymerase chain reaction (qRT-PCR).
- Proteomic analysis. Proteomic analysis includes both massive and semi-massive techniques (for proteins with medium/high and lower concentration in blood, respectively). High-performance tribrid mass spectrometry (Orbitrap Fusion Lumos, Filgen Inc., Nagoya, Japan), performed in the Centro de Regulacíón Genómica (CRG), Barcelona, according to standard methodology33 will be used for the massive analyses, while the semi-massive analyses will be based on Affimetrix arrays, targeted at specific biological facets, selected by the procedures mentioned above. Specifically, conventional techniques34 will be used to study 45 target biomarkers, including hormones and associated molecules [adrenocorticotropic hormone (ACTH), growth hormone (GH), and thyroid stimulating hormone (TSH), C-peptide, ghrelin, glucagon, insulin and leptin], inflammatory markers [chemokine expressed, and regulated on activation, normal T cell expressed and secreted (RANTES), interferon (IFN)-α2, IFN-γ, interleukin (IL)-1α, IL-1β, IL-1ra, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-15, IL-17, IP-10, monocyte chemotactic protein (MCP)-1, macrophage inflammatory protein (MIP)-1α, MIP-1β, tumor necrosis factor (TNF)-α, TNF-β, transforming growth factor (TGF)-β, and vascular endothelial growth factor (VEGF), tissue injury or remodeling markers (troponin C, desmosine, elastin, and surfactant protein D), infection markers (procalcitonin and polysaccharide-protein complex [PPC]) and endothelial dysfunction (soluble intercellular adhesion molecule [sICAM]-1). Markers that show highly discriminatory signals using these screening techniques will subsequently be determined with enzyme-linked immunosorbent assay (ELISA).
- Metabolomic analysis. Quadrupole time-of-flight mass spectrometry (Q-TOF-MS) (QSTAR XL Hybrid System, Applied Biosystems, Foster City, CA, USA) and electrospray ionization (ESI) or atmospheric pressure photoionization (APPI) will be used for metabolomic analysis. Samples will be introduced by direct infusion (DI-MS) using a low pressure pump with a 1000μL Hamilton syringe, at a flow of 5μL/min. The study will be complemented with gas chromatography (GC–MS) analysis, focusing on the more volatile metabolites. Wherever necessary, additional metabolic profiles will be obtained by mass spectrometry using ultra-high performance liquid chromatography, also using quadrupole time-of-flight analyzers (UHPLC-Q-TOF-MS). Methodology already validated in the literature will be used in all cases.35 METLIN Metabolomics Database (Scripps Research Institute, San Diego, CA, USA), FIEHN (NIH West Coast Metabolomics Center, Davis, CA, USA), HMDB (Human Metabolome Database, TMIC, Edmonton, AL, Canada) and AMIX (Bruker, Billerica, MAS, USA) metabolic databases and others will be used for the analysis of results.
Data Analysis and Statistical ProcessingClinical, functional, and conventional biological data have been collected in a single database. The platforms and statistical packages mentioned above will be used for the transcriptome, proteomic, and metabolomic analyses and the subsequent evaluation of their reciprocal interactions, along with the SPSS 18.0 (IBM Corp.) and R (GPL, free-access program), using BIOCONDUCTOR software packages (PMID: 25633503). Comparisons will be made on the basis of different dichotomous criteria, including the presence of disease, severity of disease according to functional changes (mild-moderate vs serious-very serious), presence or absence of “vulnerability” (definition given above), presence or absence of eosinophilia, presence or absence of nutritional changes, presence or absence of marked emphysema, presence or absence of bronchiectasis, male or female sex, etc. The principal components analysis (Partek Inc.) will be used for quality control and evaluation of the possible effects of homogeneity by batches. Associations between variables will be calculated using Pearson's standard and biserial correlation coefficients. A logistic regression multivariate analysis will also be performed to adjust the different covariates. A multivariate normal (MVN) model will be designed for this purpose, using the “nlme” R package (“gls” function), to assess the associations between the different clinical and biological levels. These models take all omic profiles into account simultaneously as a multivariate Y outcome. This will give a flexible variance-covariance matrix, so that the covariance depends on the major clinical variables and allows different coefficients for each selected clinical condition. Thus, the selected biological variable (Y) for each individual and clinical scenario (e.g. their phenotype) follows a multivariate normal distribution with an average vector (μ) and a covariance matrix (Σ) Y∼MVN(μ, Σ). The non-structured variance-covariance matrix is modeled using the identification of the subjects as a grouping factor. Data from the various “omics” will be integrated using multi-set canonical correlation analysis (PMID: 19377034) Finally, the results will undergo cluster analysis to define possible new disease endotypes, according to the methodology used previously by several authors of the group.10 We will rely on the human and technological resources of the Research Program on Biomedical Informatics [GRIB, property of the Hospital del Mar Institute of Medical Research (IMIM) and the UPF] for the analysis.
Population DescriptionThe recruitment phase has now been completed, with inclusion of a total of 83 healthy subjects (with two subgroups of smokers [56%] and non-smokers), 213 “non-fragile” COPD patients, and 56 more “fragile” COPD patients (Fig. 1). The lower number of patients with COPD was due mainly to the difficulty of retrieving patients from the DELICATO study, due to deaths, changes of address, or refusal to participate in this study. However, the final number of stable patients was close to 80% of the number initially planned. Furthermore, 5 controls and 36 patients were subsequently excluded when some of their blood samples were found to have deteriorated. This was mainly due to poor preservation of the sample for transcriptome analysis. Clinical data are already available from all the other subjects, derived mainly from the DELICATO study and updated at the time of inclusion in this study (Table 1). Patients are predominantly men, with good nutritional status except in the group of fragile patients (11–13% underweight), and show functional changes characteristic of their lung disease. On the basis of severity classified according to FEV1, non-fragile patients were distributed equally between mild-moderate and severe-very severe,2 while in the fragile group, a greater proportion were severe-very severe. With regard to the most recent GOLD classification, non-fragile patients showed a similar distribution among the 4 categories,2 while in the fragile group, more patients were classified as category D. Finally, both groups of patients showed similar percentages of eosinophilia in peripheral blood, anemia, and comorbidities (Charlson index).
General Characteristics BIOMEPOC Study Population.
| Healthy Subjects | “Non-fragile” COPD | “Fragile” COPD | |
|---|---|---|---|
| (n=83) | (n=213) | (n=56) | |
| Age (years) | 67±9 | 69±9 | 69±9 |
| Women (%) | 36 | 33 | 35 |
| Smoking (pack-years) | 10±3 | 58±27*** | 59±25*** |
| Never smoker (%) | 42 | 1*** | 0*** |
| Former smoker (%) | 37 | 72*** | 75*** |
| Active smoker (%) | 21 | 27 | 25 |
| BMI (kg/m2) | 27.9±5.1 | 28.2±5.3 | 26.1±5.5* |
| <20kg/m2 (%) | 0 | 2.9 | 12.7*** |
| <19kg/m2 (%) | 0 | 1.4 | 10.9*** |
| FEV1(% pred.) | 97±15 | 51±19*** | 40±14***,xxx |
| FEV1 (ml) | 2893±0.775 | 1455±0.592*** | 1022±0.304***,xxx |
| FEV1/FVC (%) | 78±5 | 48±12*** | 41±11***,xxx |
| TLC (% pred) | 102±12 | 108±18** | 109±20** |
| RV (% pred) | 102±13 | 154±29*** | 175±24***,xxx |
| RV/TLC (%) | 36±10 | 55±12*** | 61±9***,xxx |
| DLCO (% pred) | 91±12 | 53±19*** | 49±19*** |
| Kco (% pred) | 94±13 | 62±20*** | 57±22*** |
| GOLD FEV1, 1–2 (%) | – | 46.7 | 23.2xxx |
| GOLD FEV1, 3–4 (%) | – | 53.3 | 76.8xxx |
| GOLD A | – | 27.1 | 5.4xxx |
| GOLD B | – | 18.2 | 1.8xxx |
| GOLD C | – | 25.7 | 21.4 |
| GOLD D | – | 29.0 | 71.4xxx |
| Eosinophils in blood | |||
| >200mm3 (%) | 18 | 52*** | 47*** |
| >300mm3 (%) | 11 | 21* | 26** |
| Anemia (%) | – | 20 | 29 |
| Charlson index | – | 3.14±1.55 | 3.54±2.41 |
% pred.: percentage predicted; DLco: diffusing capacity of the lung for carbon monoxide; FEV1: forced expiratory volume in the first second; FVC: forced vital capacity; BMI: body mass index; Kco: Krogh's index (transfer of carbon monoxide divided by the alveolar volume, DLco/AV); RV: residual volume; TLC: total lung capacity.
Anemia, according to hemoglobin value: men and postmenopausal women, ≤13g/L; premenopausal women, ≤12g/L.
Comparisons: Group of patients vs control group of healthy subjects.
Biological samples were obtained and underwent initial processing. The BIOMEPOC project is currently in the phase of evaluating the initial results of the transcriptome analysis, and some very preliminary results have been published in the form of communications to congresses.36–38
As mentioned, the project includes a number of novel aspects in the search for biomarkers. For example, text mining has been used for the generation of new hypotheses, while semi-massive and massive analytical techniques have been used to expand screening for potential markers not governed by hypothesis. Importantly, the study is not limited to an analysis of the results of each separate “omic” technique. Instead, the results will be integrated using interaction networks that can be used to interrelate these levels (multilevel analysis). This way, the complexity of the disease can be explored from a new perspective, supplemented with already available clinical, functional and conventional analytical data (often absent in the more basic biology studies).
In summary and to conclude, this BIOMEPOC project is one of the first attempts to conduct an extensive exploration of the potential biomarkers of different aspects of the clinical heterogeneity that characterizes COPD. Its main objective is to identify the biological profiles of patients (endotypes at a high level of complexity) on the basis of certain clinical correlates (phenotypes, whether previously described or not). Potentially, it can also guide the exploration of new therapeutic pathways for some of these profiles. The project is unique insofar as it combines in a single study a sufficient number of patients (especially in the context of some “omics” analyses), a complete clinical and functional evaluation conducted by respiratory specialists, the complementary use of conventional and massive screening techniques, and simultaneous exploration of various “omics” levels (transcriptomes, proteins and metabolites) and their potential interactions. We hope in this way to generate new knowledge from a multidisciplinary perspective, with the clear aim of transferring it both to the health system and, eventually, to the industry.
FundingPartially funded by SAF2014-54371 (FEDER funds), SEPAR 2015 and 2016, FUCAP 2014, and Menarini 2015.
Conflict of InterestsThe authors state that they have no conflict of interests.
Mireia Admetlló (Hospital del Mar, CIBERES, Barcelona)
Alvar Agustí (Hospital Clínic, Universitat de Barcelona, CIBERES, Barcelona)
Carlos Alvarez-Martínez (Hospital 12 de Octubre, CIBERES, Madrid)
Esther Barreiro (Hospital del Mar, IMIM, Universitat Pompeu Fabra, CIBERES, Barcelona)
Carme Casadevall (IMIM, Universitat Pompeu Fabra, CIBERES, Barcelona)
Ferran Casals (Universitat Pompeu Fabra, Barcelona)
Robert Castelo (Universitat Pompeu Fabra, Barcelona)
Ady Castro-Acosta (Hospital 12 de Octubre, CIBERES, Madrid)
Rocío Córdova (Hospital Son Espases, CIBERES, Palma)
Borja G. Cosío (Hospital Son Espases - Instituto de Investigación Sanitaria de Baleares [IdISBa], CIBERES, Palma de Mallorca)
Rosa Faner (Fundació Clínic per la Recerca Biomédica, CIBERES, Barcelona)
Laura I. Furlong (IMIM, Barcelona)
Marian García (Consorci Hospitalari de Parc Taulí, Universitat Autònoma de Barcelona, CIBERES, Sabadell)
Joaquim Gea (Hospital del Mar, IMIM, Universitat Pompeu Fabra, CIBERES, Barcelona)
José G. González-García (Hospital del Mar, Barcelona)
Carmen Hernández-Carcereny (Hospital Clínic, Universitat de Barcelona, CIBERES, Barcelona)
José Luis López-Campos (Hospital Universitario Virgen del Rocío, Universidad de Sevilla, CIBERES)
Please cite this article as: Gea J, Pascual S, Castro-Acosta A, Hernández-Carcereny C, Castelo R, Márquez-Martín E, et al. Proyecto de biomarcadores y perfiles clínicos personalizados en la enfermedad pulmonar obstructiva crónica (proyecto BIOMEPOC). Arch Bronconeumol. 2019;55:93–99.