MicroRNA (miR) was implicated in the tumorigenesis of many types of cancer, but no study was conducted on the exact role of miR-133 in lung cancer.
MethodsWe have identified miR-133 as a putative regulator of FOXQ1 expression, and investigated the potential involvement of miR-133 in the migration and invasion of lung cancer cells, as well as the underlying molecular mechanism.
ResultsMiR-133 directly targeted and down-regulated FOXQ1 expression, which in turn reduced TGF-β level. MiR-133 was down-regulated in lung cancer cell lines A549 and HCC827, and its re-expression significantly inhibited the migration and invasion of the lung cancer cells. Further investigation revealed that this inhibition was caused by reversing the epithelial–mesenchymal transition, evidenced by miR-133 induced elevation of epithelial marker E-cadherin, and reduction of mesenchymal marker Vimentin.
ConclusionsOur study is the first to identify miR-133 as a biomarker for lung cancer. It functions to down-regulate FOXQ1, and inhibit epithelial–mesenchymal transition, which antagonizes lung cancer tumorigenesis. Therefore our data support the role of miR-133 as a potential molecular therapeutic tool in treating lung cancer.
El microARN (miR) se ha relacionado con la génesis tumoral en muchos tipos de cáncer, pero ningún estudio ha examinado el rol exacto del miR-133 en el cáncer de pulmón.
MétodosIdentificamos el miR-133 como posible regulador de la expresión de la FOXQ1 e investigamos la posible implicación del miR-133 en la migración y la invasión de células de cáncer de pulmón, y el mecanismo molecular subyacente.
ResultadosEl miR-133 se dirigió directamente y redujo la expresión de la FOXQ1, que a su vez redujo la concentración de TGF-β. El miR-133 disminuyó en líneas celulares de cáncer de pulmón A549 y HCC827, y su reexpresión inhibió significativamente la migración y la invasión de células de cáncer de pulmón. Investigaciones subsiguientes revelaron que dicha inhibición estaba provocada por una inversión de la transición epitelio-mesenquimatosa, constatada por una elevación del marcador epitelial E-cadherina inducida por el miR-133 y una reducción del marcador vimentina.
ConclusionesNuestro estudio es el primero que ha identificado el miR-133 como biomarcador del cáncer de pulmón. Su función es reducir la FOXQ1 e inhibir la transición epitelio-mesenquimatosa, la cual antagoniza la génesis tumoral en el cáncer de pulmón. Por consiguiente, nuestros datos respaldan el papel del miR-133 como posible instrumento terapéutico molecular en el tratamiento del cáncer de pulmón.
Lung cancer is the leading worldwide cause of cancer-related death.1 Epithelial–mesenchymal transition (EMT) plays an essential role in the metastasis of human cancers, including lung cancer.2 During EMT, epithelial cells acquire the mesenchymal phenotype and detach from the primary tumor, spreading through the bloodstream to invade a secondary tissue, causing metastasis.3 EMT involves both phenotypic changes and molecular reprogramming. During the transition, polarized immotile epithelial cells change their morphology to motile mesenchymal cells, assuming increased motility and invasion abilities.4 This transition is also characterized by a reduction in epithelial markers that enhance cell-cell contact, such as E-cadherin, and an increase in mesenchymal markers, such as vimentin and N-cadherin.5
Various signaling pathways have been reported to trigger EMT. Forkhead transcription factor FOXQ1 was found to induce EMT in breast cancer.6 It was also reported to mediate the crosstalk between TGF-β and Wnt signaling pathways in colorectal cancer.7 In lung cancer particularly, FOXQ1 expression contributes to poor prognosis, promotes EMT, and increases chemo-sensitivity.8,9
MicroRNAs (miRNAs or miRs) are small non-coding RNA molecules, usually 20–25 nucleotides long. They recognize specific complementary sequences commonly found in the 3′-untranslated region (UTR) of target mRNAs, either repressing translation or degrading the target mRNAs.10,11 MiRNAs are involved in the EMT of many types of cancers, including lung cancer, altering the expression of oncogenes or tumor suppressor genes.12 For example, miR-19 was found to trigger EMT and promote migration and invasion of lung cancer cell lines A549 and HCC827.13 In contrast, other miRNAs inhibit this process. One such miRNA is miR-129, which was reported to antagonize EMT and metastasis in non-small cell lung cancer by down-regulating MCRS1.14 Similarly, low expression of miR-30c was also found to negatively affect the EMT of lung cancer.15 However, the exact role of miR-133 in regulating EMT in lung cancer remains unclear.
In our current study, we aimed to investigate whether miR-133 is involved in the regulation of EMT in lung cancer cells, and the possible underlying mechanisms of this phenomenon. We found that miR-133 was down-regulated in both A549 and HCC827 human lung cancer cell lines, and miR-133 directly targeted and down-regulated FOXQ1 expression. More importantly, miR-133 expression reversed EMT features, inhibiting the migratory and invasive abilities of lung cancer cells. MiR-133 could serve as a novel molecular therapeutic tool for reversing EMT in lung cancer.
Materials and MethodsCell lines, Antibodies and MicroRNAsThe human normal lung cell lines CCD-19Lu and MRC-9, and human lung cancer cell lines A549 and HCC827 were obtained from the American Type Culture Collection (ATCC). Cells were maintained in complete medium with high-glucose RPMI-DMEM (Hyclone; GE Healthcare) and 10% fetal bovine serum (FBS; Gibco), at 37°C in a humidified atmosphere containing 5% CO2. Antibodies against FOXQ1 (ab194564), TGF-β (ab9758), total Smad2/3 (ab63672), phospho-Smad2/3 (ab63399), E-cadherin (ab76055), N-cadherin (ab18203), vimentin (ab92547), and GAPDH (ab181602) were all purchased from Abcam. Hsa-miR-133 mirVana miRNA mimic (MC10029) and mirVana miRNA negative control (4464061) were both purchased from Life Technologies. Transfection into cell lines was performed following manufacturer's instructions. After transfection, cells were incubated for another 48h before being subjected to other experimental assays.
Western BlotCells were seeded in 6-well plates at a density of 1×106 cells/well with 2ml complete DMEM medium. Twenty-four hours after transfection of either hsa-miR-133 or negative control, total protein was extracted using RIPA buffer (50mM Tris–HCl, pH 7.4, 150mM NaCl, 1mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and protease inhibitors). The protein concentration was determined using the BCA assay. Equal quantities of proteins were separated by SDS-PAGE and transferred to nitrocellulose membrane using a semi-dry transfer unit (Bio-Rad Laboratories). The membrane was then immersed in blocking buffer (PBS, 0.1% Tween-20) with 5% nonfat milk for 20min and then incubated with appropriate primary antibodies overnight at 4°C. After washing with blocking buffer, blots were incubated with HRP-conjugated secondary antibody (Life Technologies) at room temperature for 1h, washed again with blocking buffer, and visualized using Luminata Forte Western HRP Substrate (EMD Millipore).
Wound-Healing AssayA total of 1×105 cells transfected with miRNA negative control or hsa-miR-133 were seeded into a 6-well plate after transfection. A linear wound was carefully made with a 100μl sterile pipette tip across the confluent cell monolayer, and the cell debris was removed by washing with PBS. The migrated distance of the growing edge on wounded monolayers was measured at 24h after being wounded, and migration rate was expressed as a percentage relative to respective controls.
Cell invasion AssayCell invasion assay was performed in a 24-well plate with 8μm pore size chamber inserts (BD Biosciences). A total of 1×105 cells transfected with miRNA negative control or hsa-miR-133 were placed into the upper chamber per well with the Matrigel-coated membrane, diluted with serum-free culture medium. The lower compartment was filled with 500μl of medium containing 10% FBS to attract cells. The cells were incubated at 37°C in a 5% CO2 humidified incubator for 24h, followed by exposure to 20μM 5-ethynyl-2′-deoxyuridine (EdU) for an additional 4h at 37°C. Membrane inserts were removed from the plate and stained using the ENU kit (Invitrogen). The cells were counted under six random microscopic fields for each well, and invasion rate was expressed as a percentage relative to respective controls.
Cell Attachment and Detachment AssayThe cells were seeded in a 24-well plate (1×105 cells/well). Briefly, for attachment assay, after 1h incubation, non-attached cells were washed twice with PBS, and the attached cells were counted after trypsinization. The attachment rate was expressed as percentage of cell numbers of the attached cells to total cells, and normalized to respective control. For the detachment assay, the cells were detached with 0.05% trypsin for 3min and counted after the seeded cells were incubated 24h. The remaining attached cells were further trypsinized with 0.25% trypsin and counted. Cells detachment rate was expressed as a percentage of the detached cells to total cells, and normalized to respective control.
Statistical AnalysisAll values were presented as mean±standard error mean (SEM). A 2-tailed Student's t-test was used to establish significant differences between groups. Data were determined to be statistically different when P was <.05.
ResultsMiR-133 is Down-regulated in Lung Cancer Cell LinesWe found that the miR-133 sequence has a putative binding site on the 3′-UTR of FOXQ1 mRNA, using the microRNA.org resource. Moreover using RT-PCR, upon examination of its expression in two human lung cancer cell lines A549 and HCC827, we found miR-133 levels were significantly lower than in two normal lung cell lines CCD-19Lu and MRC-9. MiR-133 levels in CCD-19Lu and MRC-9 were similar, whereas in A549 it was down-regulated to approximately 30% of the normal cell lines, and in HCC827 to about 40% of the normal cell lines. These results suggested that, compared with normal human lung cells, miR-133 was significantly down-regulated in lung cancer cell lines.
MiR-133 Directly Targets 3′-UTR of FOXQ1We next validated the putative binding site of miR-133 on the 3′-UTR of FOXQ1, using a luciferase reporter assay. The sequences from wild type (WT-Luc) 3′-UTR of FOXQ1 mRNA and its mutated version (mut-Luc) were fused to the downstream of luciferase ORF, respectively. The two luciferase constructs were then separately transfected into both A549 and HCC827 cells, together with either miRNA negative control or miR-133 mimic. The luciferase reporter activity was then assessed. As a result, miR-133 dramatically reduced WT-Luc activity to approximately 30% of the control transfected experiments, consistently in both A549 and HCC827 cell lines. On the other hand, activity of mut-Luc was largely unaffected in either miR-133 or control miRNA transfected cells. The complementary sequences on FOXQ1 3′-UTR were then a direct target of miR-133.
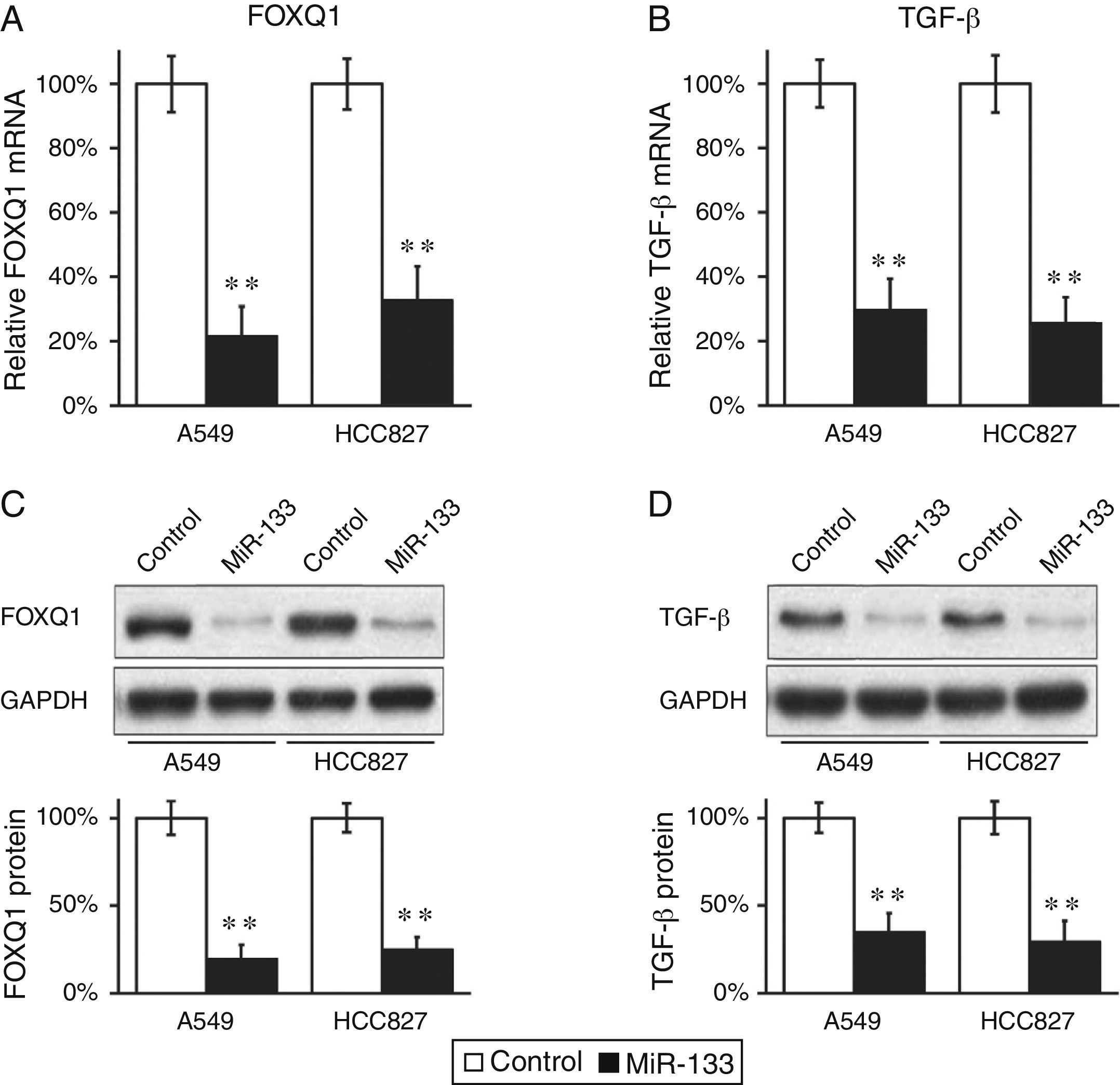
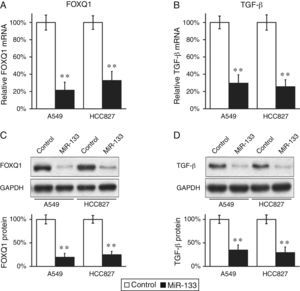
MiR-133 Down-regulates Expressions of FOXQ1 and TGF-β in Lung Cancer Cell LinesWe continued to determine whether FOXQ1 was indeed an in vivo target of miR-133. MiR-133 and a negative miRNA control were transfected into both A549 and HCC827 cells, which were then subjected to RT-PCR to examine FOXQ1 levels in mRNA. Levels of miR-133 in both A549 and HCC827 cells were indeed greatly increased upon transfection. Compared with control miRNA transfections, miR-133 significantly reduced FOXQ1 messenger levels in both lung cancer cell lines examined (Fig. 1A), clearly indicating that FOXQ1 mRNA is a bona fide target of miR-133. Using anti-FOXQ1antibody, we confirmed that its protein level was also markedly reduced in miR-133, but not in control transfected cells (Fig. 1C).
FOXQ1 is down-regulated by transfection of miR-133 into lung cancer cell lines.
(A and B) Relative FOXQ1 (A) and TGF-β (B) mRNA levels in A549 and HCC827 cell lines, after transfection with either miRNA negative control or miR-133. (C and D) FOXQ1 (C) and TGF-β (D) protein levels in A549 and HCC827 cell lines, after transfection with either miRNA negative control or miR-133. GAPDH was used as a loading control. Quantification of FOXQ1 and TGF-β protein expressions normalized to GAPDH was also shown in the lower panels. Values were mean±standard error mean (SEM) of three independent experiments. **P<.01 to respective control.
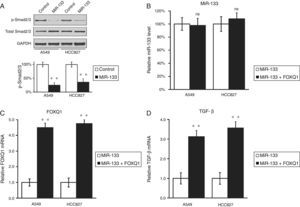
Other authors have reported that FOXQ1 could promote the expression of TGF-β,16 and was involved in the crosstalk between TGF-β and Wnt signaling pathways in the progression of colorectal cancer.7 We were curious to investigate whether a similar correlation also existed between these two genes in lung cancer. Interestingly, down-regulation of FOXQ1 by miR-133 transfection also greatly reduced TGF-β expressions in both lung cancer cell lines (Fig. 1B and D), suggesting that FOXQ1 and TGF-β are both likely to be involved in tumorigenesis of lung cancer. More importantly, we found that in both lung cancer cell lines A549 and HCC827, miR-133 expression resulted in a significant inhibition of TGF-β pathway, as indicated by reduced phosphorylation of Smad2/3 (Fig. 2A), both of which are downstream effectors of the TGF-β pathway.17 Of note, we further confirmed that the regulation of TGF-β pathway by miR-133 was indeed mediated through FOXQ1, since in a co-transfection experiment using both miR-133 and a plasmid expressing FOXQ1 without its 3′-UTR, TGF-β level was also upregulated upon FOXQ1 expression, even in the presence of miR-133 (Fig. 2B–D).
MiR-133 inhibits TGF-β pathway through down-regulation of FOXQ1 in lung cancer cell lines.
(A) Total and phosphorylated Smad2/3 protein levels in A549 and HCC827 cell lines, after transfection with either miRNA negative control or miR-133. GAPDH was used as loading control. Quantification of p-Smad2/3 expression normalized to GAPDH was also shown in the lower panels. (B–D) Relative levels of miR-133 (B), FOXQ1 mRNA (C) and TGF-β mRNA (D) in A549 and HCC827 cell lines, after transfection with miR-133 alone, or co-transfection with miR-133 and plasmid expressing FOXQ1 in the absence of its 3′-UTR. Values were mean±SEM of three independent experiments. ns not significant to respective control; **P<.01 to respective control.
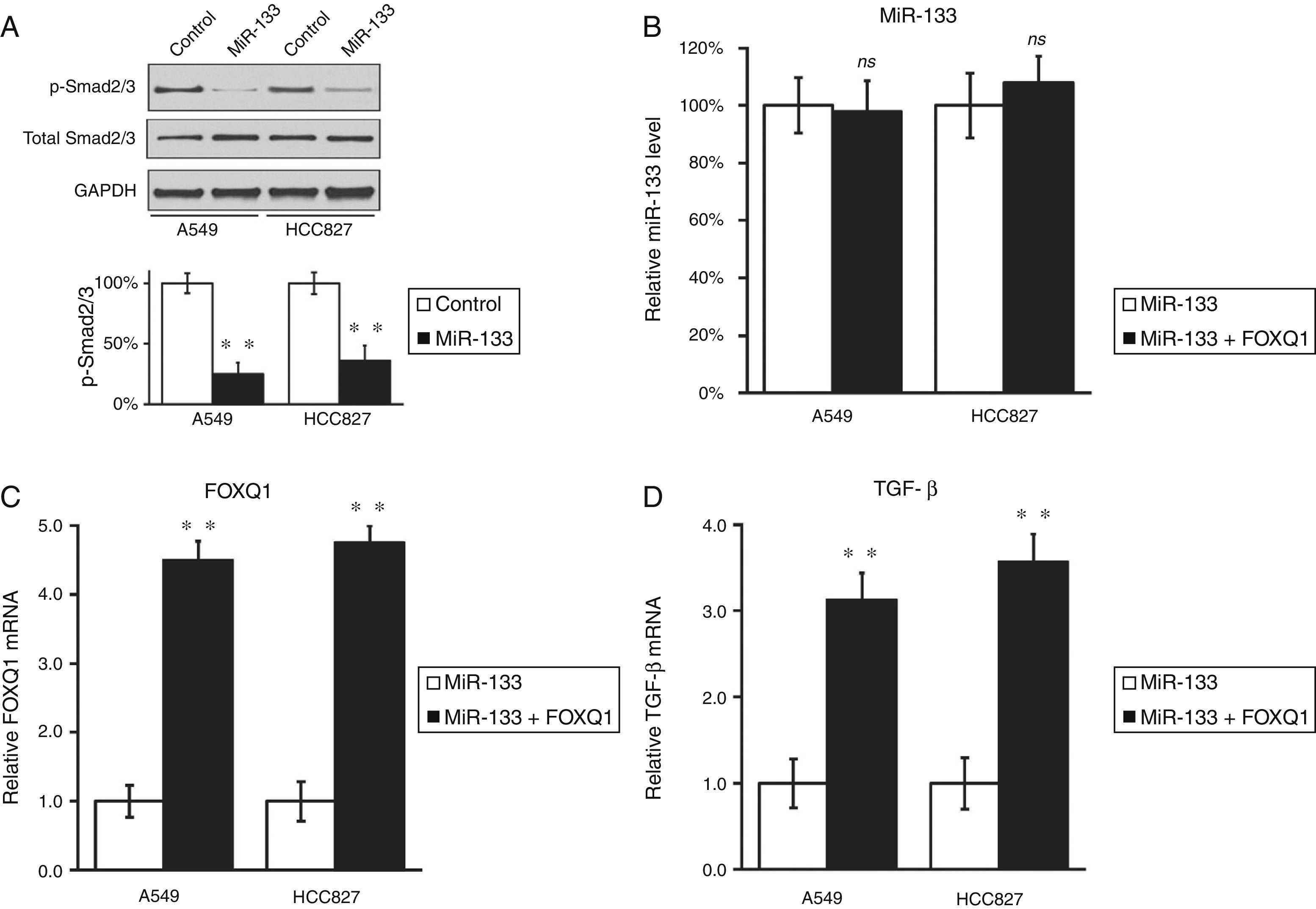
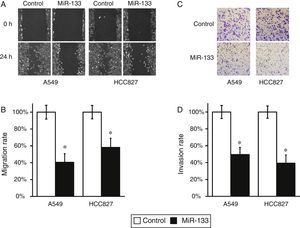
FOXQ1 is a marker for metastasis and poor prognosis of lung cancer,8,9 so introducing miR-133 to down-regulate FOXQ1 should inhibit the migration and invasion of lung cancer cells. To test this hypothesis, we performed wound-healing and cell invasion assays in both miR-133 and control miRNA-transfected A549 and HCC827 cells. As expected, both cell lines exhibited significantly reduced migration rate after transfection of miR-133, compared with control experiments (Fig. 3A and B). Moreover, in a cell invasion assay, miR-133 also consistently reduced the invasion rates of both A549 and HCC827 cells, compared with control transfected experiments (Fig. 3C and D). Taken together, the above results strongly demonstrated the role of miR-133 in inhibiting lung cancer tumorigenesis.
MiR-133 expression inhibits migration and invasion of lung cancer cells.
(A) Wound-healing assay for A549 and HCC827 cell lines, after transfection with either miRNA negative control or miR-133. (B) The migration distances relative to control in wound-healing assay were measured at 24h after being wounded. (C) Cell invasion assay for A549 and HCC827 cell lines, after transfection with either miRNA negative control or miR-133. (D) The invaded cell numbers relative to control were quantified 24h after cells were seeded. Values were mean±SEM of three independent experiments. *P<.05 to respective control.
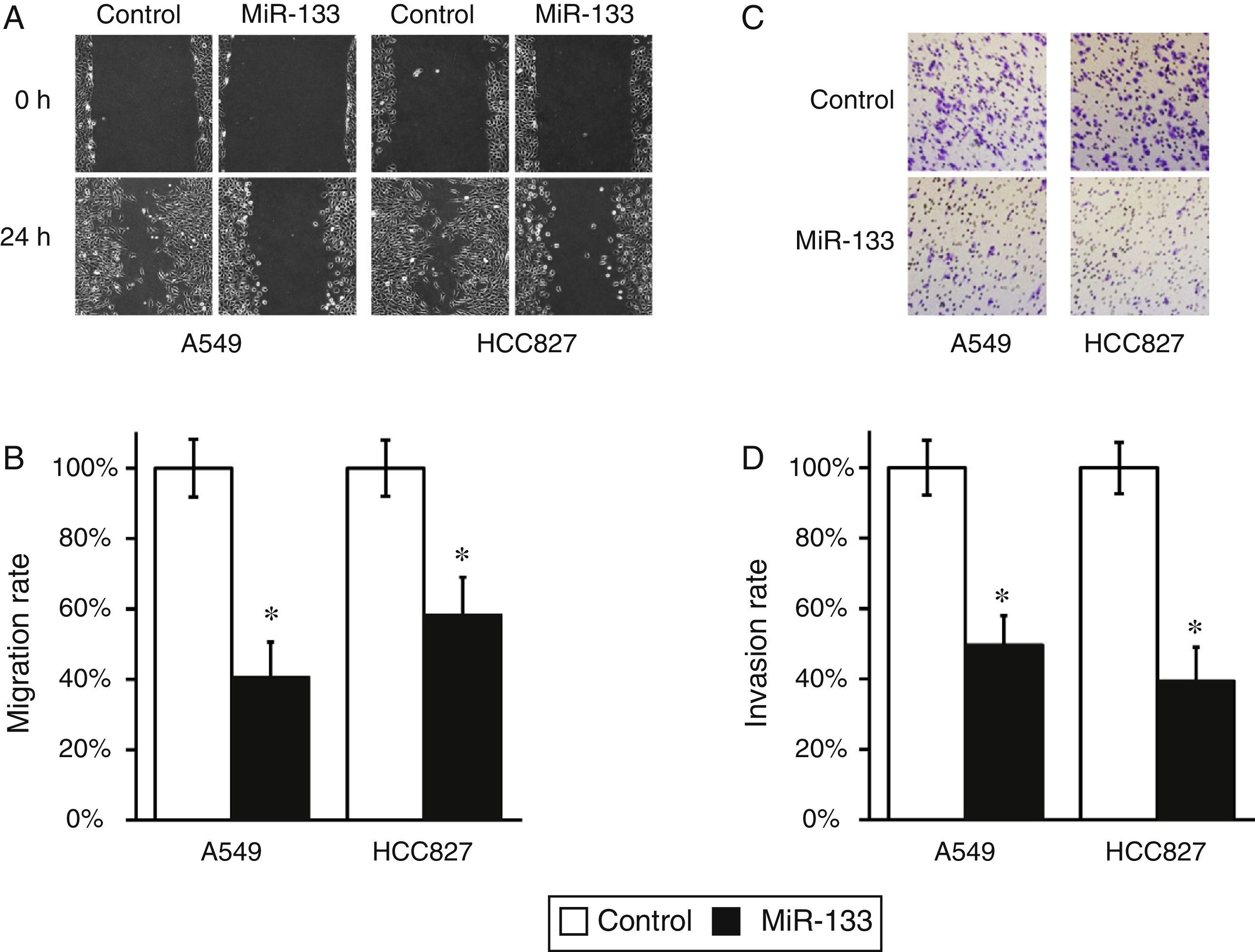
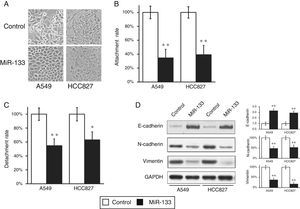
During the wound-healing and cell invasion assays, we often saw a morphological change in cells after transfection with miR-133. Both A549 and HCC827 cells initially displayed elongated and fibroblastoid morphology, whereas upon miR133 transfection both cell lines assumed rounded cell shapes (Fig. 4A), suggesting a loss of EMT phenotype. During transition, epithelial cells at the primary tumor site acquire the mesenchymal phenotype, resulting in enhanced invasion and metastasis, spreading through the blood stream to reach secondary tissues. To confirm this observation, we performed cell attachment and detachment assays in the two lung cell lines, after transfection with either miRNA negative control or miR-133. As shown in Fig. 4B and C, miR-133 significantly reduced attachment and detachment rates of both A549 and HCC827 cells, compared with control transfected cells. These results confirmed the loss of EMT features at the cell morphological level.
MiR-133 expression inhibits epithelial–mesenchymal transition (EMT) of lung cancer cells.
(A) Cell morphology was observed for A549 and HCC827 cell lines, after transfection with either miRNA negative control or miR-133. (B and C) Cell attachment (B) and detachment (C) assays were assessed in A549 and HCC827 cell lines, after transfection with either miRNA negative control or miR-133. (D) Expression of EMT phenotypic protein markers were analyzed in A549 and HCC827 cell lines, after transfection with either miRNA negative control or miR-133. Quantification of protein expressions normalized to GAPDH was also shown in the right panels. Values were mean±SEM of three independent experiments. *P<.05, **P<.01 to respective control.
At the molecular level, EMT is characterized by reduced epithelial marker E-cadherin and increased mesenchymal marker vimentin. Accordingly, we went on to assess if the same shift in EMT phenomena could also be observed at the molecular level. As expected, miR-133 increased E-cadherin expression, but reduced vimentin and N-cadherin expressions, compared with control transfected cells (Fig. 4D), which clearly demonstrated a reverse of EMT phenotype at the protein level.
DiscussionIncreasing numbers of studies have demonstrated the essential roles of different miRNAs in the tumorigenesis of lung cancer. An understanding of the abnormal expression patterns of miRNAs in lung cancer cells, as well as the underlying molecular programs, could contribute to the development of new clinical therapies in the treatment of lung cancer. In this study, we have demonstrated that miR-133 was down-regulated in two human lung cancer cell lines A549 and HCC827, compared to normal lung cell lines CCD-19Lu and MRC-9. Interestingly, in silico analysis showed that miR-133 has a putative binding site on the 3′-UTR of FOXQ1 mRNA. Using a luciferase reporter assay, we confirmed that the 3′-UTR sequence was a direct target of miR-133. Furthermore, by reintroducing miR-133 expression into the A549 and HCC827 lung cancer cell lines, we found both mRNA and protein levels of FOXQ1 were consistently reduced, suggesting that FOXQ1 expression was indeed repressed by miR-133 in vivo. On the other hand, expression of TGF-β was also down-regulated by miR-133 transfection. This finding was consistent with a previous report which suggests that it is a downstream effector of FOXQ1.16 Moreover, miR-133 expression significantly reduced the migration and invasion of lung cancer cells, probably through EMT inhibition, as evidenced by a prominent elevation of epithelial marker E-cadherin and reduction of mesenchymal markers N-cadherin and vimentin.
Recent studies have highlighted the important roles of miRNAs in the EMT of cancer cells. For example, the miR-134/487b/665 cluster regulates TGF-β mediated EMT by targeting membrane-associated guanylate kinase inverted 2 in lung cancer cells.18 Additionally, miR-125b/489 was found to regulate EMT pathway in breast cancer.19,20 Down-regulation of miR-200c was found to be responsible for acquired resistance to EGFR inhibitors by manifesting EMT features.21 Moreover, miR-216a/217 was found to induce EMT by targeting PTEN and Smad7 in liver cancer.22
Emerging studies have highlighted the role of miR-133 in several types of human cancers. It functions as a tumor suppressor by targeting IGF-1R in hepatocellular carcinoma.23 In colorectal cancer, ectopic expression of miR-133 significantly suppressed cancer growth both in vitro and in vivo, by targeting E3-ubiquitin protein ligase, thereby increasing p53 expression.24 Moreover, miR-133 was also widely down-regulated in several gastric cancer cell lines, where its re-expression was found to repress CDC42-PAK signaling pathway and lead to inhibited cancer cell migration and invasion.25 Quite a few recent studies have consistently reported that miR-133 was also able to directly target epidermal growth factor receptor (EGFR) in prostate cancer,26 bladder cancer,27 gastric cancer,28 and glioblastoma multiforme,29 inhibiting proliferation, migration, and invasion of these cancer cells. Together, all the above studies suggest that miR-133 plays a critical and complex role in regulating progression of different human cancers by targeting different signaling pathways.
In conclusion, our study is the first report implicating miR-133 in the EMT of lung cancer. As EMT is commonly associated with aggressiveness of cancer cells, including migration and invasion, inhibiting or reversing this transition could lead to new therapeutic approaches in the treatment of cancer. In this context, we found that miR-133 could reverse the EMT of lung cancer cell lines A549 and HCC827, which may explain why miR-133 could significantly inhibit the migration and invasion of the lung cancer cells. Our results suggest that miR-133, by inhibiting the epithelial–mesenchymal transition, may have a role as a potential molecular therapeutic tool in the treatment of lung cancer.
Author ContributionsAll authors participated in the design and interpretation of the studies, analysis of the data and review of the manuscript. Bo Xiao, Huazhen Liu, and Zeyun Gu conducted the experiments and Bo Xiao and Cheng Ji wrote the manuscript.
FundingThis work was supported by National Natural Science Foundation of China (31300969), Natural Science Foundation of Jiangsu Province (BK20130302) and China Postdoctoral Science Foundation (2013M540461 and 2013M541717).
Conflict of InterestThe authors declare no conflict of interest.
Please cite this article as: Xiao B, Liu H, Gu Z, Ji C. La expresión de microARN-133 inhibe la transición epitelio-mesenquimatosa en las células del cáncer de pulmón apuntando directamente al FOXQ1. Arch Bronconeumol. 2016;52:505–511.