In some recent issues, such as that of Belmonte et al.1 published in the International Journal of COPD, have incorporated of the Mmalton allele-specific genotyping assay in the diagnostic algorithm of alpha-1 antitrypsin deficiency (AATD) to allow the clinical characterization of Mmalton individuals.
Current laboratory tests for AATD involve the determination of a combination of alpha-1 antitrypsin (AAT) serum levels, AAT phenotyping by isoelectric focusing, and an allele specific genotyping assay to detect the most prevalent, S and Z, deficiency alleles.2 However, rare variants can only be detected by more complex techniques, such as the use of allelic specific probes or sequencing of the SERPINA1 gene, which are not available in all routine laboratories.
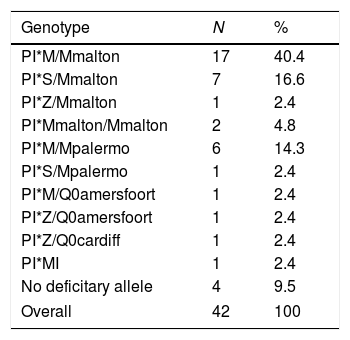
A study published by Martinez Bugallo et al.3 emphasizes that the Mmalton variant in the third most frequent variant deficiency in our island, after S and Z alleles, and with a higher prevalence than that described in the Iberian Peninsula. Forty two patients with AAT values <100mg/dL and with an inconclusive result in the genotype for PI*S and PI*Z underwent complete sequencing of the SERPINA1 gene. Of the 42 patients studied, at least one infrequent deficient allele was detected in 90.4% of the cases (38 patients). The most common deficient variant was Mmalton allele (64.2%), followed by Mpalermo allele (16.6%), both caused by the F52del mutation (Table 1).
Genotypes of Subjects with Rare Deficiency Alleles.
| Genotype | N | % |
|---|---|---|
| PI*M/Mmalton | 17 | 40.4 |
| PI*S/Mmalton | 7 | 16.6 |
| PI*Z/Mmalton | 1 | 2.4 |
| PI*Mmalton/Mmalton | 2 | 4.8 |
| PI*M/Mpalermo | 6 | 14.3 |
| PI*S/Mpalermo | 1 | 2.4 |
| PI*M/Q0amersfoort | 1 | 2.4 |
| PI*Z/Q0amersfoort | 1 | 2.4 |
| PI*Z/Q0cardiff | 1 | 2.4 |
| PI*MI | 1 | 2.4 |
| No deficitary allele | 4 | 9.5 |
| Overall | 42 | 100 |
The Mmalton and Mpalermo are two rare variants characterized by an F52del (c.226_228delTTC) mutation. While the Mmalton allele must have derived from the normal M2 allele, Mpalermo derives from the normal M1V.4,5
In our population, Mpalermo represents 1 in 5 individuals with the F52del mutation, and although the use of specific probes for the detection of this mutation seems to be a good diagnostic strategy, it should be used as screening, since that in our opinion it is necessary to perform the complete sequencing of SERPINA1 in all cases to make a more accurate diagnosis of these variants, being necessary to confirm the presence of the base allele M2 or M1V in cis in these patients.