MicroRNA-7 (miR-7) has a suppressive role in lung cancer and alterations in its DNA methylation may contribute to tumorigenesis. As COPD patients with emphysema have a higher risk of lung cancer than other COPD phenotypes, we compared the miR-7 methylation status among smoker subjects and patients with various COPD phenotypes to identify its main determinants.
Methods30 smoker subjects without airflow limitation and 136 COPD patients without evidence of cancer were recruited in a prospective study. Clinical and functional characteristics were assessed and patients were classified into: frequent exacerbator, emphysema, chronic bronchitis and asthma COPD overlap (ACO). DNA collected from buccal epithelial samples was isolated and bisulfite modified. miR-7 methylation status was evaluated by quantitative methylation-specific polymerase chain reaction (qMSP).
ResultsmiR-7 Methylated levels were higher in COPD patients than in smokers without airflow limitation (23.7±12.4 vs. 18.5±8.8%, p=0.018). Among COPD patients, those with emphysema had higher values of methylated miR-7 (27.1±10.2%) than those with exacerbator (19.4±9.9%, p=0.004), chronic bronchitis (17.3±9.0%, p=0.002) or ACO phenotypes (16.0±7.2%, p=0.010). After adjusting for clinical parameters, differences between emphysematous patients and those with other phenotypes were retained. In COPD patients, advanced age, mild-moderate airflow limitation, reduced diffusing capacity and increased functional residual capacity were identified as independent predictors of methylated miR-7 levels.
ConclusionThe increase of miR-7 methylation levels experienced by COPD patients occurs mainly at the expense of the emphysema phenotype, which might contribute to explain the higher incidence of lung cancer in these patients.
El microRNA-7 (miR-7) tiene un papel supresor en el cáncer de pulmón, y las alteraciones en la metilación de su DNA podrían contribuir a la tumorogénesis. Como los pacientes con EPOC y enfisema presentan un mayor riesgo de sufrir cáncer de pulmón frente a otros fenotipos de EPOC, comparamos la metilación de miR-7 entre los pacientes fumadores y los pacientes con varios fenotipos de EPOC para identificar sus factores determinantes principales.
MétodosSe reclutaron para un estudio prospectivo 30 sujetos fumadores sin restricciones en el flujo aéreo y 136 pacientes con EPOC sin evidencia de cáncer. Se valoraron las características clínicas y funcionales y se clasificaron a los pacientes en: exacerbaciones frecuentes, enfisema, bronquitis crónica y solapamiento de asma y EPOC (ACO, por sus siglas en inglés). Se recogió ADN a partir de muestras de epitelio bucal, se aisló y se modificó con bisulfito. El estado de metilación del miR-7 se evaluó mediante la reacción cuantitativa en cadena de la polimerasa específica de la metilación (qMPS por sus siglas en inglés).
ResultadosLos niveles de metilación del miR-7 fueron más altos en los pacientes con EPOC que en los fumadores sin restricciones en el flujo aéreo (23,7±12,4 frente a 18,5±8,8%, p=0,018). Entre los pacientes con EPOC, aquellos con enfisema presentaban valores más altos de miR-7 metilado (27,1±10,2%) que aquellos con exacerbaciones (19,4±9,9%, p=0,004), bronquitis crónica (17,3±9,0%, p=0,002) o los fenotipos ACO (16,0±7,2%, p=0,010). Tras ajustar los resultados a los parámetros clínicos, las diferencias entre los pacientes enfisematosos y aquellos con otros fenotipos permanecieron. En los pacientes con EPOC, se identificaron como predictores independientes de los niveles de metilación del miR-7 a: la edad avanzada, limitación al flujo aéreo leve-moderada, capacidad de difusión reducida y capacidad residual funcional aumentada.
ConclusiónEl aumento de los niveles de metilación del miR-7 que experimentan los pacientes con EPOC ocurre principalmente a expensas del fenotipo con enfisema, lo que podría contribuir a explicar la mayor incidencia de cáncer de pulmón en estos pacientes.
Chronic obstructive pulmonary disease (COPD) is a leading cause of morbidity and mortality worldwide. Epidemiological studies have consistently demonstrated that the presence of COPD increases the incidence of lung cancer and its mortality.1 In fact, COPD could increase the risk of lung cancer two- to six-fold, regardless of smoking habits.1
The presence of specific proteinases produced by immune and stromal cells, inflammatory cytokines, cell cycle deregulation and several genetic and epigenetic changes have been implicated as potential pathogenic mechanisms linking COPD and lung cancer.2 In recent years, there has been much evidence suggesting that microRNAs (miRNAs), a class of small non-coding RNAs involved in post-transcriptional gene repression, have an emerging role in cancer, acting mainly as tumor suppressors.3 It has also been suggested that miRNAs, which regulate cellular processes such as proliferation, cell invasion, migration, adaptation to hypoxia, apoptosis, or angiogenesis, may be involved in lung tumorigenesis in COPD patients.4 In fact, the expression of several miRNAs in induced sputum, bronchoalveolar lavage or lung tissue has shown to be lower in currently-smoking patients with COPD than in healthy subjects.5
Recently, microRNA-7 (miR-7) has been reported to act as a suppressor in several types of cancer, including non-small-cell lung cancer (NSCLC),6 and its reduced levels have been associated with increased tumorigenicity.7–9 Moreover, in patients with lung cancer, low miR-7 expression levels have been related with poorer tumor differentiation, advanced pathological T-factor, higher incidence of lymph node metastasis and advanced p-TNM stage.6 In contrast, enhanced miR-7 expression suppresses the growth of lung cancer and promotes cell apoptosis.10 Nevertheless, the mechanism underlying the downregulation of miR-7 in patients at risk for lung cancer remains largely unknown. Although mutations in the miR-7 promoter region have been described in some subjects,11 epigenetic changes through DNA methylation could be the main mechanism to suppress miR-7 function by inhibiting its expression.12,13 In fact, many methylated miRNA genes have been found in patients with lung cancer,12 and previous reported studies from our group have shown that the methylation of miR-7 is an event related to tumor progression in lung cancer cells that is also present in early stages in NSCLC patients.14 miR-7 methylation modulates the redox response through its direct target gene MAFG, conferring cell protection against free radicals.15
An important aspect to consider in the COPD/lung cancer relationship is that COPD is a very heterogeneous disease; several studies have demonstrated that lung cancer risk and mortality vary between the different disease phenotypes.16,17 Regarding other COPD phenotypes, patients with emphysema have a higher risk of both small- and non-small-cell lung cancers.18 Despite this, little progress has been made in the identification of mechanistic links between COPD phenotypes and lung cancer. In particular, no information is available on whether the different COPD phenotypes are associated with different miR-7 methylation patterns, which could justify different risks for developing lung cancer. Therefore, our objective was to compare the methylation status of miR-7 among smoker subjects and patients with several COPD phenotypes in order to identify the main determinants of miR-7 methylation levels in these patients.
MethodsStudy subjectsConsecutive sampling was used to recruit 136 COPD patients with the next inclusion criteria: postbronchodilator FEV1/FVC<0.7 and>20 pack-years; stable condition at inclusion, with no infection or exacerbation for at least 2 months; and optimal medical therapy according to GesEPOC recommendations19 for at least 8 weeks with no change. As control group, 30 smoker subjects (>20 pack-years) without evidence of airflow limitation (FEV1/FVC>0.7 and >lower limit of normal) were consecutively recruited from the lung cancer screening program of our center. The study was approved by the institutional ethics committee and each participant gave written informed consent (HULP-PI 2109). A more detailed description of the methods is provided in the supplementary material.
ProceduresClinical and functional evaluationAnthropometric characteristics, smoking habit, comorbidities and current treatment were recorded. In COPD patients, dyspnea was graded using the modified Medical Research Council (mMRC) score, and exacerbations requiring treatment with antibiotics, oral corticosteroids and/or hospitalization in the year prior to the study were also recorded.
Arterial blood gas values breathing room air, spirometric parameters, lung volumes and diffusing capacity for carbon monoxide were measured according to current recommendations. Global Lung Initiative (GLI) predicted values were used for spirometry20 and those of the European Coal and Steel Community21 for lung volumes and diffusing capacity. Airflow limitation severity and COPD risk groups were stratified according to the GOLD statement.22
According to the GesEPOC criteria,19 patients were classified in the next phenotypes: frequent exacerbator, chronic bronchitis, emphysema and asthma COPD overlap (ACO).
Analysis of miR-7 DNA methylationThe DNA from the 166 buccal epithelial swab samples was eluted with phosphate-buffered saline (PBS) and digested by proteinase K (Invitrogen, Carlsbad, CA), followed by extraction with phenol/chloroform and used for methylation analysis of the specific CpG positions after bisulfite modification of DNA as previously reported.14,15 The sodium bisulfited samples were used for quantitative methylation-specific polymerase chain reaction (qMSP) by designing two probes (6-carboxyfluorescein [FAM] and VIC®), to specifically recognize methylated or unmethylated CpG positions, making it possible to measure the percentage of methylated DNA in a unique PCR reaction. We used the following primer/probe set for methylated reaction: F: 5′-GGGTGGGGTTTTTTAAGAATC-3′; R: 5′-ACATTCTCCTCCTTCGATCG-3′; Probe: 5′-FAM-ACCCCTCTTCGTTCTCGAT-3′; and for unmethylation: F: 5′-GGGGTGGGGTTTTTTAAGAATT-3′; R: 5′-ATAACATTCTCCTCCTTCAATCA-3′; Probe: 5′-VIC-ACCCCTCTTCATTCTCAAT-3′, using the following settings: 50°C–2min; 95°C–15min; and 40 cycles of 94°C–50 seg, 60°C–1min. All assays were performed in duplicate using the QuantiTect Multiplex PCR Kit (Qiagen, USA) and the HT7900 Applied Biosystems. Each set of modified DNA included DNA from healthy donors as a negative control and DNA methylated in vitro with Sss I methylase (New England Biolabs, Beverly, MA) to be used as a positive control. The percentage of methylation of each sample was calculated as previously reported.15
Statistical analysisData are summarized as mean±SD for continuous variables, while frequencies (percentages) are used for categorical variables. The normality of the distribution of variables was tested using the Kolmogorov–Smirnov and Shapiro–Wilk tests. Differences between study groups were analyzed using analysis of variance with post hoc comparisons by the Bonferroni test or by the chi-square test. The effect of the possible confounding factors was assessed using generalized linear models. We constructed a multivariate model, including COPD phenotype as fixed factor and gender, age, BMI, smoking status, airflow limitation severity, GOLD risk group, and treatment with inhaled corticosteroids as covariates. The relationships between methylated miR-7 levels and clinical/functional characteristics of COPD patients were determined using Pearson's linear bivariate correlation. Gender, age, smoking habit, airflow limitation severity and significantly related variables were then used in a multiple linear regression analysis to identify independent determinants of methylated miR-7 levels. All analyses were performed using SPSS 13.0 software. A p<0.05 was accepted as the minimum level of statistical significance.
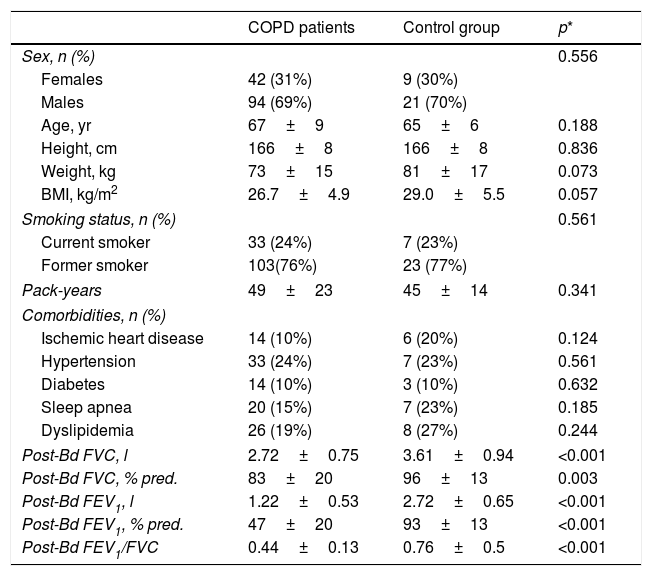
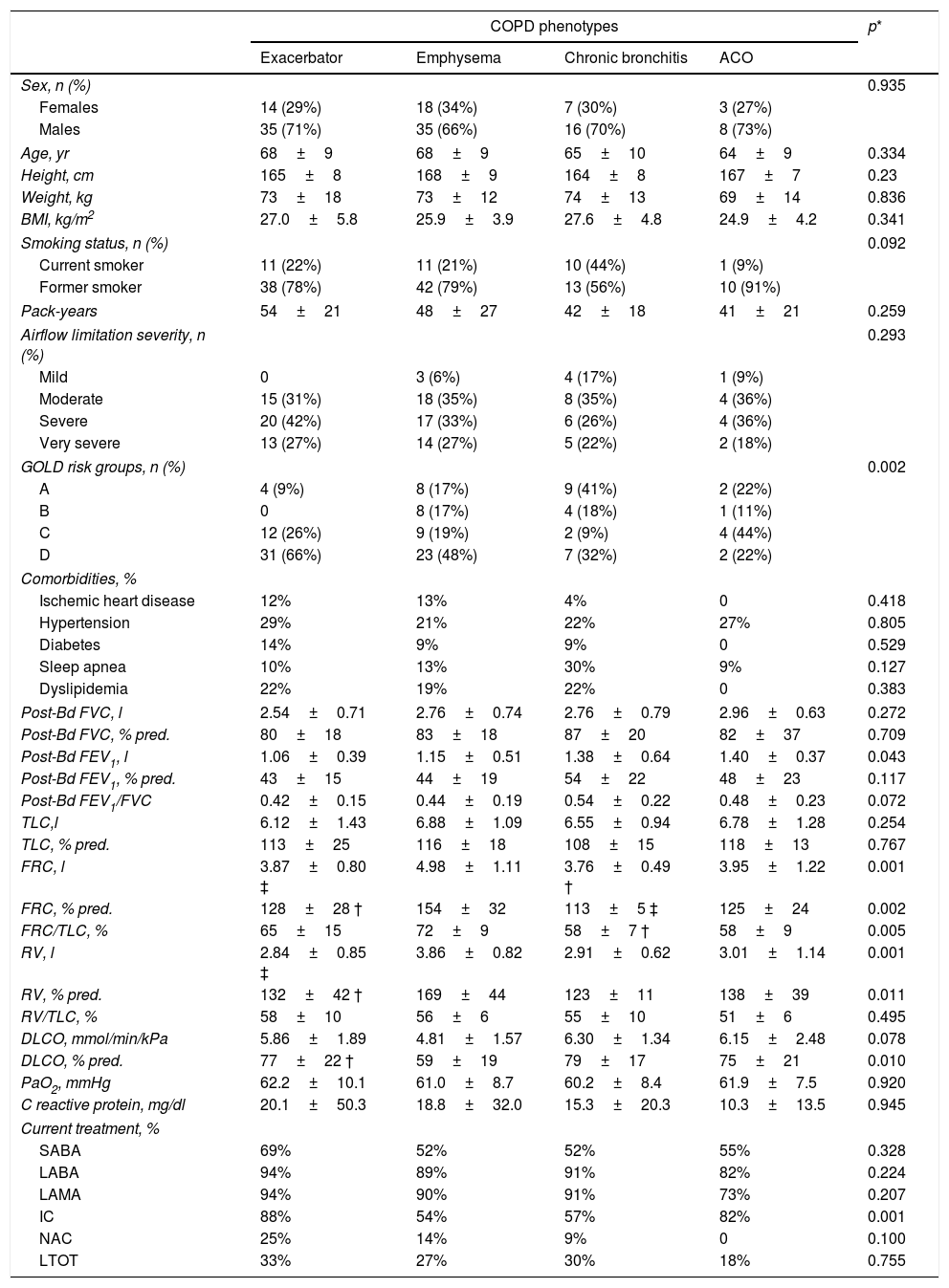
ResultsTable 1 shows the general characteristics of the control subjects and COPD patients, verifying that both groups are homogeneous in anthropometric and smoking characteristics as well as in the presence of comorbidities. Forty-nine COPD patients (36%) presented the exacerbator phenotype, 53 (39%) emphysematous phenotype, 23 (17%) chronic bronchitis phenotype, and the remaining 11 (8%) had ACO phenotype. Comparisons of anthropometric, clinical and functional characteristics of the four COPD phenotypes are shown in Table 2. As expected, differences in static lung volumes and diffusing capacity were found between the emphysematous group and other COPD patients. Moreover, treatment with inhaled corticosteroids was more frequent in patients with ACO. No other relevant differences were found between the COPD patients.
General characteristics of the study subjects.
| COPD patients | Control group | p* | |
|---|---|---|---|
| Sex, n (%) | 0.556 | ||
| Females | 42 (31%) | 9 (30%) | |
| Males | 94 (69%) | 21 (70%) | |
| Age, yr | 67±9 | 65±6 | 0.188 |
| Height, cm | 166±8 | 166±8 | 0.836 |
| Weight, kg | 73±15 | 81±17 | 0.073 |
| BMI, kg/m2 | 26.7±4.9 | 29.0±5.5 | 0.057 |
| Smoking status, n (%) | 0.561 | ||
| Current smoker | 33 (24%) | 7 (23%) | |
| Former smoker | 103(76%) | 23 (77%) | |
| Pack-years | 49±23 | 45±14 | 0.341 |
| Comorbidities, n (%) | |||
| Ischemic heart disease | 14 (10%) | 6 (20%) | 0.124 |
| Hypertension | 33 (24%) | 7 (23%) | 0.561 |
| Diabetes | 14 (10%) | 3 (10%) | 0.632 |
| Sleep apnea | 20 (15%) | 7 (23%) | 0.185 |
| Dyslipidemia | 26 (19%) | 8 (27%) | 0.244 |
| Post-Bd FVC, l | 2.72±0.75 | 3.61±0.94 | <0.001 |
| Post-Bd FVC, % pred. | 83±20 | 96±13 | 0.003 |
| Post-Bd FEV1, l | 1.22±0.53 | 2.72±0.65 | <0.001 |
| Post-Bd FEV1, % pred. | 47±20 | 93±13 | <0.001 |
| Post-Bd FEV1/FVC | 0.44±0.13 | 0.76±0.5 | <0.001 |
Definition of abbreviations: BMI=body mass index; FVC=forced vital capacity; FEV1=forced expiratory volume at 1 second.
Clinical and functional characteristics of the COPD patients according their clinical phenotypes.
| COPD phenotypes | p* | ||||
|---|---|---|---|---|---|
| Exacerbator | Emphysema | Chronic bronchitis | ACO | ||
| Sex, n (%) | 0.935 | ||||
| Females | 14 (29%) | 18 (34%) | 7 (30%) | 3 (27%) | |
| Males | 35 (71%) | 35 (66%) | 16 (70%) | 8 (73%) | |
| Age, yr | 68±9 | 68±9 | 65±10 | 64±9 | 0.334 |
| Height, cm | 165±8 | 168±9 | 164±8 | 167±7 | 0.23 |
| Weight, kg | 73±18 | 73±12 | 74±13 | 69±14 | 0.836 |
| BMI, kg/m2 | 27.0±5.8 | 25.9±3.9 | 27.6±4.8 | 24.9±4.2 | 0.341 |
| Smoking status, n (%) | 0.092 | ||||
| Current smoker | 11 (22%) | 11 (21%) | 10 (44%) | 1 (9%) | |
| Former smoker | 38 (78%) | 42 (79%) | 13 (56%) | 10 (91%) | |
| Pack-years | 54±21 | 48±27 | 42±18 | 41±21 | 0.259 |
| Airflow limitation severity, n (%) | 0.293 | ||||
| Mild | 0 | 3 (6%) | 4 (17%) | 1 (9%) | |
| Moderate | 15 (31%) | 18 (35%) | 8 (35%) | 4 (36%) | |
| Severe | 20 (42%) | 17 (33%) | 6 (26%) | 4 (36%) | |
| Very severe | 13 (27%) | 14 (27%) | 5 (22%) | 2 (18%) | |
| GOLD risk groups, n (%) | 0.002 | ||||
| A | 4 (9%) | 8 (17%) | 9 (41%) | 2 (22%) | |
| B | 0 | 8 (17%) | 4 (18%) | 1 (11%) | |
| C | 12 (26%) | 9 (19%) | 2 (9%) | 4 (44%) | |
| D | 31 (66%) | 23 (48%) | 7 (32%) | 2 (22%) | |
| Comorbidities, % | |||||
| Ischemic heart disease | 12% | 13% | 4% | 0 | 0.418 |
| Hypertension | 29% | 21% | 22% | 27% | 0.805 |
| Diabetes | 14% | 9% | 9% | 0 | 0.529 |
| Sleep apnea | 10% | 13% | 30% | 9% | 0.127 |
| Dyslipidemia | 22% | 19% | 22% | 0 | 0.383 |
| Post-Bd FVC, l | 2.54±0.71 | 2.76±0.74 | 2.76±0.79 | 2.96±0.63 | 0.272 |
| Post-Bd FVC, % pred. | 80±18 | 83±18 | 87±20 | 82±37 | 0.709 |
| Post-Bd FEV1, l | 1.06±0.39 | 1.15±0.51 | 1.38±0.64 | 1.40±0.37 | 0.043 |
| Post-Bd FEV1, % pred. | 43±15 | 44±19 | 54±22 | 48±23 | 0.117 |
| Post-Bd FEV1/FVC | 0.42±0.15 | 0.44±0.19 | 0.54±0.22 | 0.48±0.23 | 0.072 |
| TLC,l | 6.12±1.43 | 6.88±1.09 | 6.55±0.94 | 6.78±1.28 | 0.254 |
| TLC, % pred. | 113±25 | 116±18 | 108±15 | 118±13 | 0.767 |
| FRC, l | 3.87±0.80 ‡ | 4.98±1.11 | 3.76±0.49 † | 3.95±1.22 | 0.001 |
| FRC, % pred. | 128±28 † | 154±32 | 113±5 ‡ | 125±24 | 0.002 |
| FRC/TLC, % | 65±15 | 72±9 | 58±7 † | 58±9 | 0.005 |
| RV, l | 2.84±0.85 ‡ | 3.86±0.82 | 2.91±0.62 | 3.01±1.14 | 0.001 |
| RV, % pred. | 132±42 † | 169±44 | 123±11 | 138±39 | 0.011 |
| RV/TLC, % | 58±10 | 56±6 | 55±10 | 51±6 | 0.495 |
| DLCO, mmol/min/kPa | 5.86±1.89 | 4.81±1.57 | 6.30±1.34 | 6.15±2.48 | 0.078 |
| DLCO, % pred. | 77±22 † | 59±19 | 79±17 | 75±21 | 0.010 |
| PaO2, mmHg | 62.2±10.1 | 61.0±8.7 | 60.2±8.4 | 61.9±7.5 | 0.920 |
| C reactive protein, mg/dl | 20.1±50.3 | 18.8±32.0 | 15.3±20.3 | 10.3±13.5 | 0.945 |
| Current treatment, % | |||||
| SABA | 69% | 52% | 52% | 55% | 0.328 |
| LABA | 94% | 89% | 91% | 82% | 0.224 |
| LAMA | 94% | 90% | 91% | 73% | 0.207 |
| IC | 88% | 54% | 57% | 82% | 0.001 |
| NAC | 25% | 14% | 9% | 0 | 0.100 |
| LTOT | 33% | 27% | 30% | 18% | 0.755 |
Definition of abbreviations: ACO=asthma-COPD overlap; BMI=body mass index; GOLD=Global Initiative for Chronic Obstructive Lung Disease; FVC=forced vital capacity; FEV1=forced expiratory volume at 1 second; TLC=total lung capacity; FRC=functional residual capacity; RV=residual volume; DLCO=diffusion capacity for carbon monoxide; PaO2=arterial pressure of oxygen; SABA=short-acting beta-adrenergic drugs; LABA=long-acting beta-adrenergic drugs; LAMA=long-acting muscarinic antagonists; IC=inhaled corticosteroids; NAC=N-acetylcysteine; LTOT=long-term oxygen-therapy
Methylation levels of miR-7 were higher in patients with COPD than in smokers without airflow limitation (23.7±12.4 vs. 18.5±8.8%, p=0.018). This difference was retained after adjusting for sex, age, BMI and smoking habit (mean±standard error of mean [SEM], 22.1±0.9 vs. 18.3±1.7%, p=0.042).
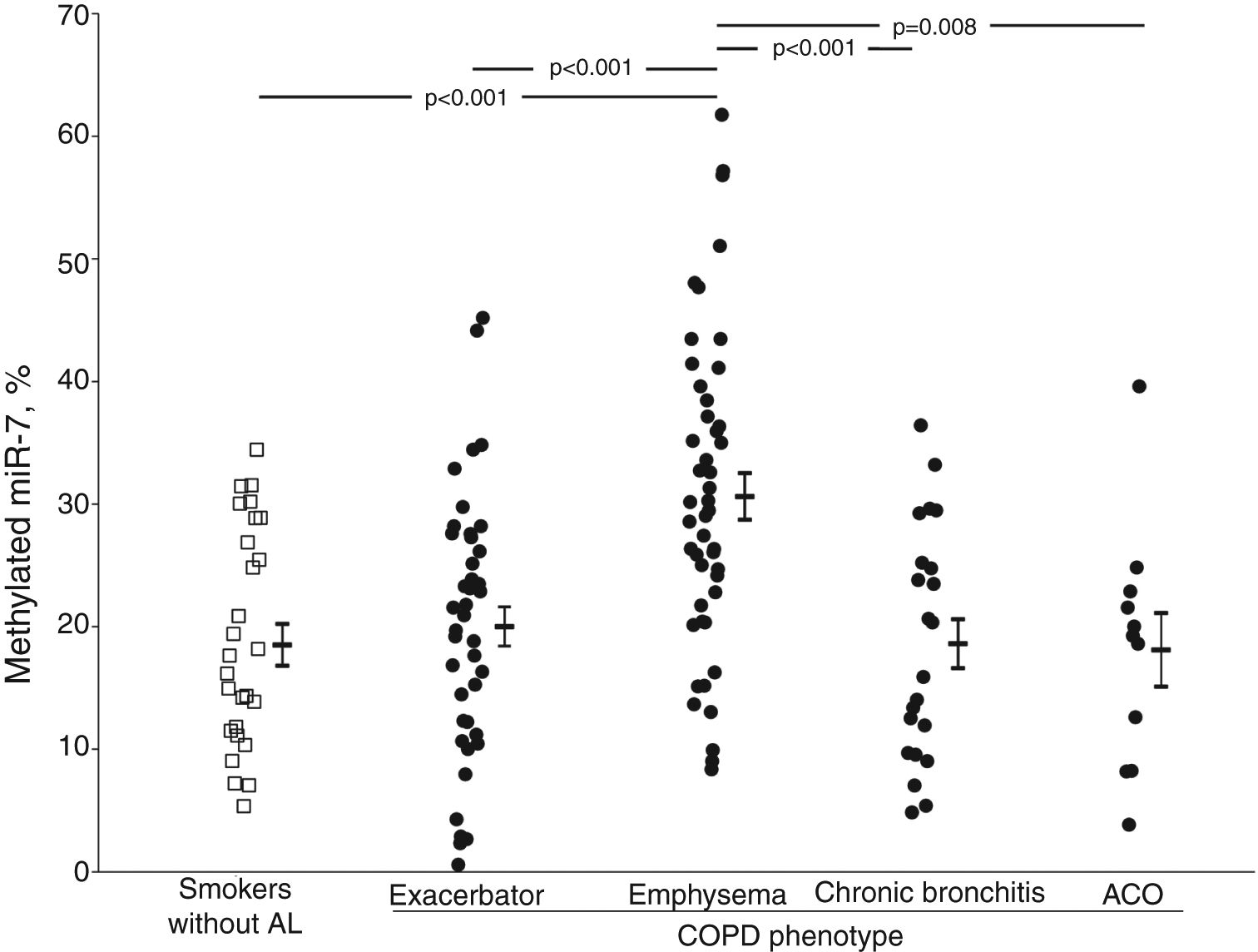
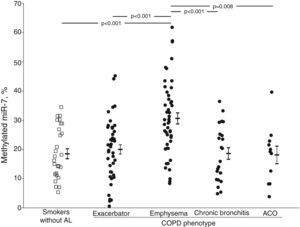
The crude comparison of methylated miR-7 levels showed that COPD patients with emphysema had higher levels (30.6±12.8%) than the smoker subjects without airflow limitation (p<0.001) and the exacerbator (20.0±10.6%, p<0.001), chronic bronchitis (18.6±9.5%, p<0.001) and ACO phenotype groups (18.1±9.9%, p=0.008) (Fig. 1). After adjusting for gender, age, BMI, smoking status, airflow limitation severity, GOLD risk group and treatment with inhaled corticosteroids, phenotype dependence was retained. Adjusted mean of methylated miR-7 levels continued to be higher in the emphysematous phenotype (mean±SEM, 26.4±1.5%) than in the other COPD phenotypes (19.3±1.5%, p=0.008 in exacerbator phenotype; 18.7±2.1%, p=0.027 in chronic bronchitis phenotype; and 16.1±3.4%, p=0.048 in ACO phenotype).
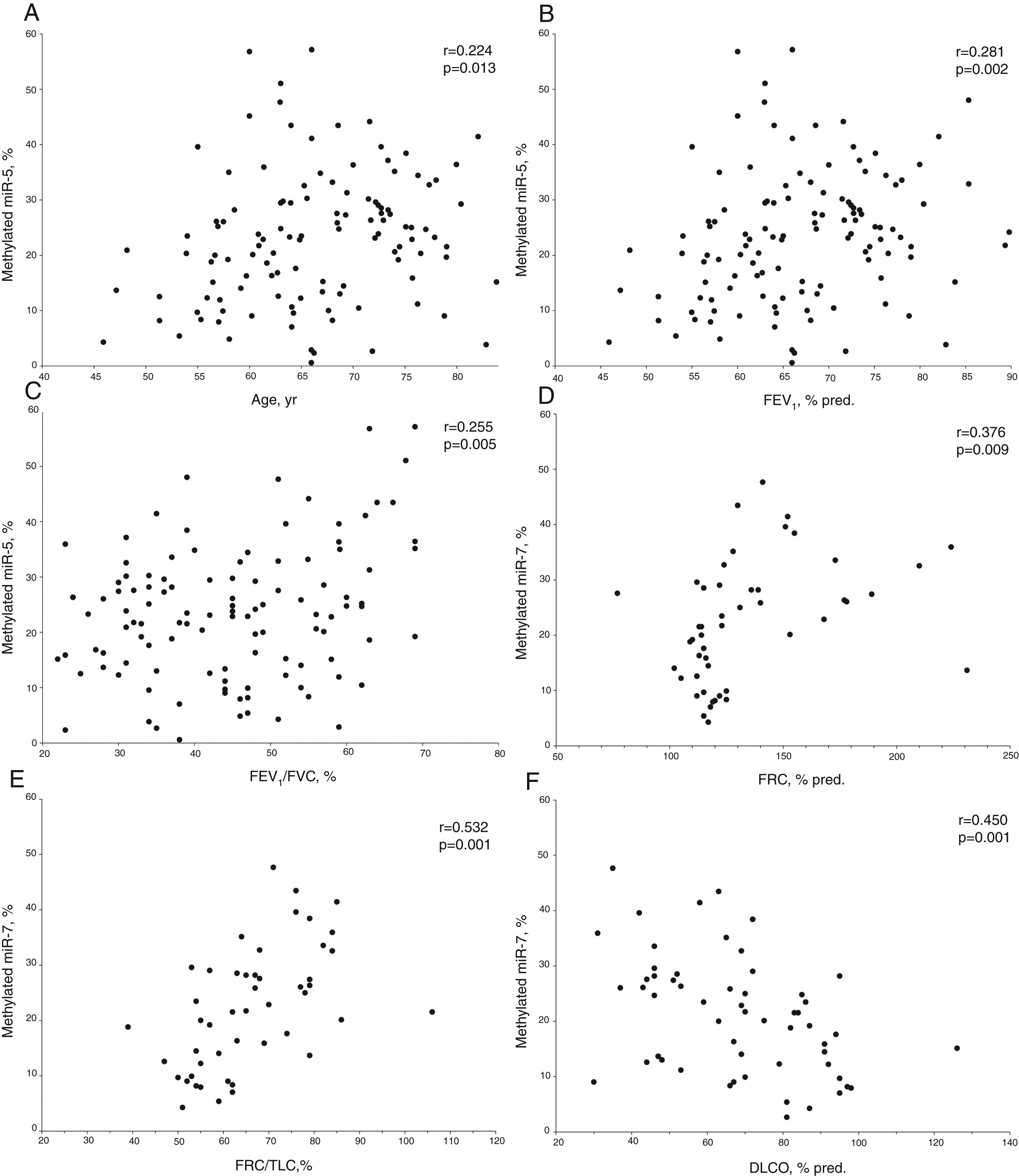
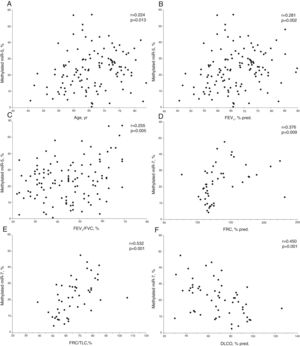
In COPD patients, methylated miR-7 levels directly correlated with age (r=0.224, p=0.013), forced expiratory volume at 1 second (FEV1) (r=0.281, p=0.002), FEV1/forced vital capacity (FVC) ratio (r=0.255, p=0.005), functional residual capacity (FRC) (r=0.376, p=0.009) and FRC/total lung capacity (TLC) ratio (r=0.532, p<0.001), while an inverse relationship was found with diffusing capacity (r=−0.450, p<0.001) (Table S1 and Fig. 2). Interestingly, no differences were found in methylated levels of miR-7 according to gender (24.2±13.2% in males versus 22.5±10.6% in females, p=0.491), smoking habit (23.2±12.9% in current smokers versus 23.8±12.4% in former smokers, p=0.804), or use of inhaled corticosteroids (22.5±11.5% in patients with IC versus 25.1±13.4% in patients without IC, p=0.278).
Relationship between age (A), forced expiratory volume at 1 second (FEV1) (B), FEV1/forced vital capacity (FVC) ratio (C), functional residual capacity (FRC) (D), FRC/total lung capacity (TLC) ratio (E) and diffusing capacity (DLCO) (F) with methylated miR-7 levels in COPD patients. r=Pearson's correlation coefficient.
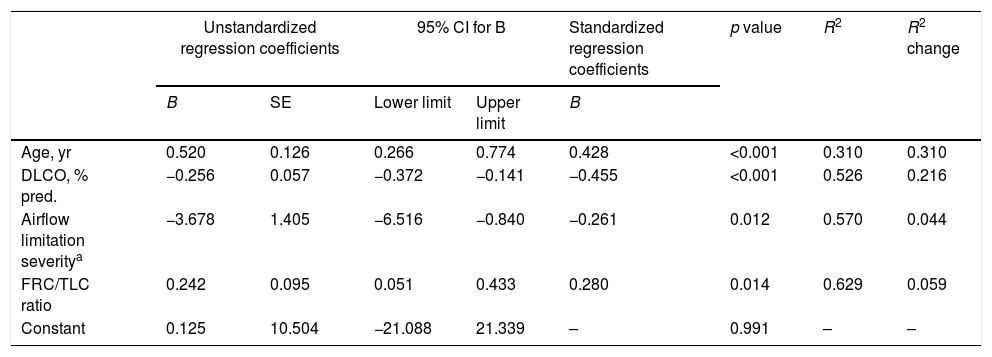
The stepwise multiple regression model for methylated miR-7 levels of COPD patients only retained age, airflow limitation severity, diffusing capacity and FRC/TLC ratio as independent variables (Table 3). The model that included these four variables accounted for 62.9% of the explained variance in methylated miR-7 found in these patients.
Independent predictors of methylated miR-7 level of COPD patients in a multivariate linear regression analysis.
| Unstandardized regression coefficients | 95% CI for B | Standardized regression coefficients | p value | R2 | R2 change | |||
|---|---|---|---|---|---|---|---|---|
| B | SE | Lower limit | Upper limit | B | ||||
| Age, yr | 0.520 | 0.126 | 0.266 | 0.774 | 0.428 | <0.001 | 0.310 | 0.310 |
| DLCO, % pred. | −0.256 | 0.057 | −0.372 | −0.141 | −0.455 | <0.001 | 0.526 | 0.216 |
| Airflow limitation severitya | −3.678 | 1.405 | −6.516 | −0.840 | −0.261 | 0.012 | 0.570 | 0.044 |
| FRC/TLC ratio | 0.242 | 0.095 | 0.051 | 0.433 | 0.280 | 0.014 | 0.629 | 0.059 |
| Constant | 0.125 | 10.504 | −21.088 | 21.339 | – | 0.991 | – | – |
Definition of abbreviations: DLCO=diffusing capacity for carbon monoxide; FRC=functional residual capacity; TLC=total lung capacity; CI=confidence interval.
The most relevant findings of our study are the demonstration that the increase miR-7 methylation levels found in COPD patients with respect smoker subjects without airflow limitation occurs mainly at the expense of patients with emphysema, and that advanced age, mild or moderate airflow limitation, static hyperinflation and decreased lung diffusing capacity are the main determinants of methylation levels in COPD patients. Considering that lower miR-7 levels have been linked to the development of lung cancer,7 these findings might contribute to explaining the increased risk of lung cancer identified in patients with emphysema versus other COPD types.18,23 In fact, a recent analysis of the BODE cohort has shown that the presence of emphysema in COPD patients is independently associated with lung cancer diagnosis.23 In turn, another analysis of the same group showed that the presence of reduced DLCO in patients with COPD has a similar impact on the risk of lung cancer as CT-determined emphysema.24
In recent years, multiple miRNAs have been identified as having suppressive roles,6 and their dysregulation has been associated with lung cancer development.25 Epigenetic alterations through DNA methylation may inhibit miRNA expression, contributing to the onset of cancer. Moreover, DNA methylation has been implicated in the altered expression of numerous oncogenes and tumor suppressors in lung cancer,2 and many methylated miRNA genes have been found in NSCLC.12 Particularly, miR-7 has been well documented as a promising tumor suppressor by targeting multiple oncogenes,6 and previous studies in our laboratory have shown a reduction in miR-7 expression in chemotherapy-resistant subtypes of H23 lung cancer cells through hypermethylation of its regulatory region14 as well as a negative correlation between the percentage of methylation and expression levels of miR-7 in tumors from 22 NSCLC patients.15 Several reports demonstrate that miR-7 down-regulates epidermal growth factor receptor (EGFR)-mediated oncogenesis via inactivation of ERK, Akt and MAPK signaling pathways, inducing cell cycle arrest and cell death.6,26,27 In contrast, aberrant expression and function of miR-7 has been associated with tumorigenesis,9 promoting cell proliferation, anchorage-independent cell growth, and tumor formation of lung cancer cells.8
There was a striking coincidence between the variables independently related with miR-7 methylation in our patients and the classical risk factors for lung cancer in COPD patients. Thus, the methylation of miR-7 was more intense in older patients and those with a lower degree of airflow limitation. In this context, it is well known that COPD patients have a higher risk for developing cancer as they age.23,24 At the same time, and while being somewhat controversial, de Torres et al. observed an inverse relationship between the incidence of lung cancer and COPD severity.28 Hypermethylation patterns could be the cause of, or a result of, altered lung function. Current information does not allow discriminate whether it is only the expression of a greater susceptibility to tobacco exposition or might also have some pathogenic role in the airflow limitation progression.29 Furthermore, COPD patients may also suffer from other conditions related to severity of airflow limitation, such as hypoxemia, or take drug therapies, which may be associated with differential DNA methylation. Regarding hyperinflation, in a cohort of 848 COPD patients followed for 4–5 years, we have recently reported that hyperinflation is an independent risk factor for the development of lung cancer.30 Lastly, several studies of the BODE cohort have demonstrated that a decrease in DLCO is also associated with a higher risk of lung cancer diagnosis as well as related death.24,28 In any event, similar to hyperinflation, the decrease in DLCO is a marker for the severity of CT-detected emphysema.31
Although our study is not able to define the mechanisms responsible for the miR-7 hypermethylation in the emphysema phenotype, some evidence available leads us to speculate that it could be due to smoking-induced up-regulation of matrix metalloproteinases as well as excessive inflammatory and oxidative stress responses.1,32 Emphysema is characterized by a proteinase/antiproteinase imbalance, excessive oxidative burden, increased apoptosis of lung epithelial and endothelial cells and an inflammatory cell infiltrate mainly composed of neutrophils, macrophages and lymphocytes.33 It is known that reactive oxygen species damage epithelial cells, induce a genotoxic stress capable of DNA adduct formation2 and increase miRNA methylation.34 Moreover, aberrant methylation has been described in tissues exposed to specific types of inflammation.35 Finally, matrix-degrading enzymes are essential for the development of emphysema, and the overexpression of some of these enzymes, such as matrix metalloproteinase 9, has been linked to epigenetic promoter methylation.36
Although one would expect oxidative stress induced by tobacco smoke to increase the miR-7 methylation rate, we have not found a relationship with the intensity of smoking evaluated by the pack-year rate, probably because we had recruited patients with a prolonged and intense history of exposure to smoking. In fact, our results agree with those from a large cohort of COPD-active smokers or -ex-smokers, in whom the pack-year rate was not a predictive factor for lung cancer risk.37
Another aspect to consider is the potential influence of treatment with inhaled corticosteroids, as DNA methylation is regulated by three classes of DNA methyltransferases (DNMTs) and glucocorticosteroids have an inhibitory effect on DNMT1 and DNMT3A expression.13 In fact, they regulate the expression of several miRNAs which target genes involved in regulation of metabolism, differentiation, proliferation and cell survival.38 Although the effect on methylation is more frequent in systemic steroids,20,39 budesonide may also induce changes in methylation status.40 In our opinion, the difference in the use of inhaled corticosteroids among our COPD patients does not seem to justify the differences found in miR-7 methylation levels, which are similar between patients treated or not with these drugs. In fact, the differences among COPD phenotypes are maintained after adjusting for previous treatment with inhaled corticosteroids. Perhaps the DNMT3B expression, which is also responsible for “de novo” DNA methylation in cancer,41 is not affected by inhaled corticosteroids.
Our study presents several limitations. First, it is a cross-sectional study that simply demonstrates differences in a biomarker related with the development of lung cancer among different COPD phenotypes. In order to establish that COPD patients with higher miR-7 methylation levels have a higher risk for lung cancer, it will be necessary to complete the longitudinal follow-up of this cohort in coming years. Second, there are differences in certain patient characteristics among the different COPD phenotypes, to which we have tried to adjust the comparison of miR-7 methylation levels. However, the analysis we have done does not completely rule out that one of these factors may partially contribute to the differences found. Third, miR-7 methylation levels have been evaluated systemically instead of in lung tissue due to the greater accessibility of this source, that allows a none-invasive approach. Fourth, our results are from a single-center cohort, and their verification is necessary in patients with COPD from other settings and with other characteristics, which may influence their epigenetic regulation.
In conclusion, our study demonstrates that patients with different clinical COPD phenotypes present different levels of miR-7 methylation, which is more intense in patients with the emphysema phenotype and shows an independent relationship with hyperinflation and a decline in lung diffusion capacity. This finding could provide biological plausibility to previous evidence that shows that the COPD-lung cancer relationship is heterogeneous and varies between the different phenotypes of the disease. Future studies will need to clarify whether miR-7 methylation levels could be a useful biomarker for the appropriate selection of COPD patients with an optimal risk-benefit ratio in lung cancer screening programs.
FundingThis study was supported by the ‘Fondo de Investigación Sanitaria-Instituto de Salud Carlos III’ [PI15/00186 and PI18/00050 to I.I.C., and PI13/01512, PI16/00201 an PIE15/00065 to F.G.G.]; and the European Regional Development Fund/European Social Fund FIS [FEDER/FSE, Una Manera de Hacer Europa]. MINECO funds support R.R. and O.P. contracts through and RTC-2016-5314-1 project. The funding sources did not provide any input in the development of the research and manuscript.
Conflict of interestThe authors declare no conflict of interest.