The diagnosis of microscopic lymph node metastasis in lung cancer is challenging despite the constant advances in tumor staging. The analysis of the methylation status of certain genes in lymph node samples could improve the diagnostic capability of conventional cyto-histological methods. The aim of this study was to demonstrate the feasibility of methylation studies using cytological lymph node samples.
MethodsA prospective study including 88 patients with a diagnosis or strong suspicion of non-small cell lung cancer, in which an echobronchoscopy was performed on mediastinal or hilar lymph nodes for diagnosis and/or staging purposes. DNA was extracted from cytological lymph node samples and sodium bisulfite modification was performed. Methylation studies for p16/INK4a and SHOX2 were accomplished by MS-qPCR and pyrosequencing.
ResultsThe methodology used in our study yielded optimal/good DNA quality in 90% of the cases. No differences in DNA concentration were observed with respect to the lymph node biopsied and final diagnosis. Methylation analyses using MS-qPCR and pyrosequencing were not possible in a small number of samples mainly due to low DNA concentration, inadequate purity, fragmentation and/or degradation as a consequence of bisulfite conversion.
ConclusionMethylation quantification using MS-qPCR and pyrosequencing of cytological lymph node samples obtained using echobronchoscopy is feasible if an appropriate DNA concentration is obtained, notably contributing to the identification of epigenetic biomarkers capable of improving decision making for the benefit of potentially curable lung cancer patients.
El diagnóstico de la afectación metastásica ganglionar en el cáncer de pulmón constituye un problema, a pesar de los avances en la estadificación. La determinación del estado de metilación en ganglios podría mejorar la capacidad de las técnicas citohistológicas para detectar afectación metastásica. Nuestro objetivo fue demostrar la viabilidad de realizar estudios de metilación en muestras ganglionares citológicas.
MétodosEstudio prospectivo que incluyó 88 pacientes con diagnóstico o alta sospecha de cáncer de pulmón no microcítico, en los que se realizó una punción citológica por ecobroncoscopia de adenopatías mediastínicas y/o hiliares. Se extrajo ADN a partir de muestras citológicas ganglionares y se realizó el tratamiento con bisulfito de sodio. Los estudios de metilación se realizaron por qPCR-MS y pirosecuenciación en los genes p16/INK4a y SHOX2.
ResultadosLa metodología empleada permitió obtener ADN de características óptimas/buenas en el 90% de los casos. No se observaron diferencias en la concentración de ADN respecto a la estación ganglionar ni al diagnóstico final. Los análisis por qPCR-MS y pirosecuenciación no fueron posibles en un reducido número de muestras debido a baja concentración de ADN, además de la inadecuada pureza, fragmentación y/o degradación debido al tratamiento con bisulfito de sodio.
ConclusiónLa cuantificación de la metilación por técnicas como qPCR-MS o pirosecuenciación en muestras ganglionares obtenidas por ecobroncoscopia resulta viable siempre y cuando se logre obtener una concentración adecuada de ADN, contribuyendo a la búsqueda de biomarcadores epigenéticos que mejoren la toma de decisiones en el cáncer de pulmón potencialmente curable en beneficio del paciente.
Lung cancer (LC) is currently one of the major public health problems and the most common cause of death from cancer.1 This is because most patients are identified in advanced stages of the disease, when options for curative treatments, mainly surgical, no longer exist. Recent years have seen significant advances in the development of new imaging techniques, such as positron emission tomography-computed tomography, sampling guided by endobronchial endoscopic ultrasound (EBUS) and esophageal ultrasound, and surgical exploration of the mediastinum with new mediastinoscope systems with inbuilt videocameras (mediastinal and transcervical) and video-assisted thoracoscopy, which are more profitable and improve staging accuracy.2,3 However, 30%–40% of patients without apparent lymph node involvement in surgery suffer a tumor relapse.4 Diagnosis of lymph node involvement poses many challenges and is the most important factor in the prognosis and treatment of patients with potentially resectable LC. Moreover, microscopic lymph node involvement (occult N) cannot be confirmed by routine cytohistological pathology techniques, and more accurate nodal staging is necessary.3,5 The search for strategies to improve these aspects is essential.
Advances in epigenetics, especially DNA methylation studies, have been shown to be particularly useful in LC6,7 for diagnosis, prognosis and predicting response to treatment. However, knowledge of the potential applicability of epigenetics in hilar and mediastinal staging is still limited. The aim of this study was to demonstrate the feasibility of methylation studies by assessing the quantity and quality of DNA extracted from lymph node samples obtained by endobronchial ultrasound-guided transbronchial needle aspiration (EBUS-TBNA) in patients with non-small cell lung carcinoma.
MethodsStudy SubjectsA prospective study including consecutive patients with diagnosis or high suspicion of non-small cell lung carcinoma, in which EBUS-guided needle cytology of mediastinal or hilar lymphadenopathy was performed for diagnosis and/or staging purposes. Case inclusion was determined by the multidisciplinary LC committee of the center (www.chuvi.es), according to the local protocol for diagnosis, staging and treatment of lung cancer which follows the current national and international standards and recommendations. Patients with hilar and/or mediastinal lymphadenopathy with a short axis diameter of greater than 1cm on computed tomography, or 5mm in case of pathological uptake in positron emission tomography-computed tomography, were included. Patients without an evaluable sample according to the cytologist, or an insufficient sample for further methylation studies were excluded. All procedures were performed in Bronchopleural Technical Unit of CHUVI Pneumology Service.
The study was conducted according to the clinical and ethical principles of the Spanish Government and the Declaration of Helsinki, and was approved by the Ethics Committee for Clinical Research of Galicia. Informed consent was obtained from all patients and anonymity was guaranteed.
Procedure for Endobronchial Ultrasound-Guided Transbronchial PunctureAll procedures were performed consecutively between September 2009 and December 2012. Most were ambulatory and took place in a conventional bronchoscopy room with echocardiography, blood pressure and pulse oximetry monitoring, and under conscious sedation with midazolam and fentanyl. A bronchoscope model BF-UC180F-OL8 (Olympus, Tokyo, Japan) and ultrasound equipment Aloka ProSound Alpha 5 (Hitachi-Aloka, Tokyo, Japan) were used. After endoscopic examination, the lesion was measured and punctured using a NA2015X-4022 needle (Olympus, Tokyo, Japan), advancing through the ultrasound bronchoscope canal to the airway, where the needle was extracted and inserted into the node through the tracheal and bronchial wall with ultrasound guidance. In the node the needle was mobilized and vacuum suction with syringe was performed. Depending on the immediate results obtained, one to three passes were made. The material was recovered and the sample was fixed in alcohol and immediately examined by a cytologist, who identified it as follows: normal node (predominantly lymphoid cells without atypia and/or anthracotic material); node with neoplastic infiltration (presence of neoplastic lymph node cells and cellularity); or non-evaluable sample (presence of only blood or bronchial cells). A part of the samples verified in situ as representative was resuspended in sterile saline solution and immediately frozen at −20°C for subsequent molecular studies.
In cases in which the pathology report found no neoplastic involvement, negative results were verified by thoracic surgery. Patients who did not undergo confirmation by surgery were followed through their medical records for a year. If the lymphadenopathy did not grow during the year, the result was considered a true negative.8 All doubtful cases were excluded from the study.
DNA Extraction from Lymph Node SamplesDNA was extracted using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA), adapting the protocol to the volume of each sample. DNA was eluted with 50μL of water and two aliquots were separated for further treatment with sodium bisulfite. DNA concentration was measured with NanoDrop 2000c (Thermo Scientific). The 260/280nm absorbance ratios allowed estimating their purity as optimal (ratio 1.7–2.0), good (ratio > 2.0), or presence of proteins or other contaminants (ratio < 1.7). DNA was stored at −20°C until use.
Sodium Bisulfite TreatmentMethylation analysis required treatment of DNA with sodium bisulfite. EZ DNA Methylation Direct Kit (Zymo Research, Irvine, CA, USA) was used for this purpose, following the manufacturer's recommended protocol, and eluting with 20μL. Modified DNA becomes a uracil-rich single chain, chemically similar to RNA, so its concentration was determined using the NanoDrop (Thermo Scientific), as if it were this nucleic acid. The 260/280nm ratios allowed purity to be assessed as optimal (ratio 1.8–2.2), good (ratio >2.2), or with presence of proteins or other contaminants (ratio <1.8). Treated DNA was stored at −80°C until use.
Methylation Analysis by Real-time Methylation-Specific Polymerase Chain ReactionMethylation analysis of p16/INK4a and SHOX2 genes was performed using nested methylation specific (MS) real-time PCR (qPCR), with 3μL of bisulfite-treated DNA. External primers were used in the first round to amplify the CpG island regardless of methylation status. MS-qPCR was subsequently performed by diluting the PCR product and using methylation-specific probes and primers in a StepOne equipment (Applied Biosystems). More detailed information of the sequences used are available on request.
Methylation Analysis by PyrosequencingPyrosequencing was performed for p16/INK4a and SHOX2 genes. PCR was performed with 3μL of modified DNA using primers designed with PyroMark Assay Design v2.0 (Qiagen) software. Sequencing and methylation quantification were then carried out using the pyrosequencing primer in a PyroMark Q24 (Qiagen). The CpG islands studied were the same as those analyzed by qPCR-MS. More detailed information of the sequences used are available on request.
Statistical AnalysisThe SPSS package (v19.0) was used for statistical analysis. Two-tailed tests were used, and P-values <.05 were considered significant. DNA concentration was expressed as median and interquartile range (IQR). Normal distribution and homogeneity of variance were tested with Kolmogorov–Smirnov and Levene tests, respectively. Non-parametric analysis for two independent samples was performed using Mann–Whitney U test, while Kruskal–Wallis was used for more than two independent samples. A Wilcoxon test was used to analyze differences in paired data. The variables studied were gender, tumor type, location of lymph nodes, diagnosis of lymphadenopathy, appearance of the nodal sample and concentration and purity of unmodified and sodium bisulfite-modified DNA.
Sample size was estimated using the Granmo 5.2 application (Municipal Institute of Medical Research, 1999) for paired samples, with an alpha risk of 0.05 and a beta risk of 0.20. The estimated prevalence of malignant lymph nodes was assumed to be 70%, and the assumed difference between sensitivity and negative predictive value of molecular techniques and cytohistological determinations was 15% and 10%, respectively. A minimum of 79 cases was estimated as necessary for performing the study.
ResultsPatient Characteristics and Cytological Specimens From Lymph NodesEighty-eight patients were included, yielding 170 samples from different lymph node stations obtained by EBUS-TBNA. The patients were distributed as follows: 38 patients with one sample, 29 with two samples, 13 with three samples, 5 with four samples, and 3 with five samples. Seventy-four patients (84%) were male, with a median age of 62.2 years (IQR, 51–73 years). On a proprietary color scale, 44.7% (n=76) of the cytological node samples were of turbid-clear appearance, 38.2% (n=65) were slightly bloody, and 17.1% (n=29) were very bloody.
In 86.5% of cases (n=147) punctures were performed in mediastinal lymph nodes, while 13.5% (n=23) were performed in hilar lymph nodes. The lymphadenopathy most frequently punctured was 4R (39.4%, n=67) and 7 (33.5%, n=57), followed by 4L (9.4%, n=16), 11L (7.1%, n=12), 11R (4.7%, n=8), 8 (2.4%, n=4), 2R (1.8%, n=3), 10R (1.2%, n=2) and 12R (0.6%, n=1). According to the definitive lymph node diagnosis, 48.2% (n=82) were benign (non-metastatic), while the remaining 51.8% (n=88) were malignant (metastatic). Among these, 61.4% (n=54) were adenocarcinoma, 31.8% (n=28) squamous cell carcinoma and 6.8% (n=6) large cell carcinoma.
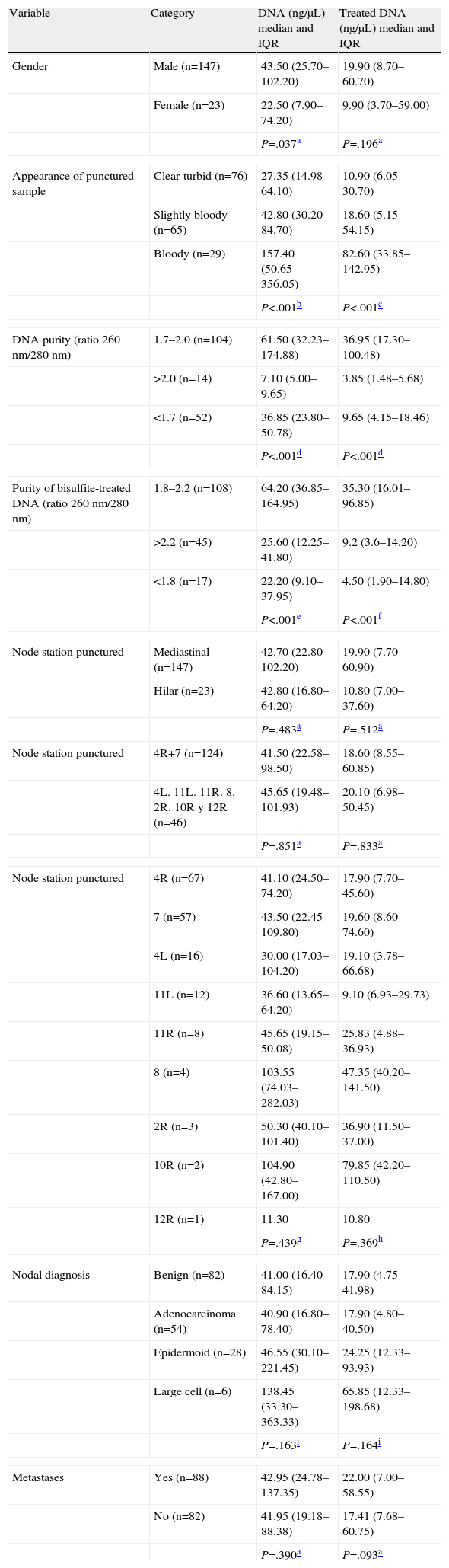
Characteristics of Deoxyribonucleic Acid Extracted From Lymph Node Cytological SpecimensA wide range of DNA concentrations was obtained, from 4.10 to 1103.00ng/μL (median: 42.75ng/μL; IQR: 22.48–100.13ng/μL). In order to study DNA characteristics, concentrations were related to epidemiological and clinical parameters (Table 1). Males presented higher median values, with significant differences compared to women (P=.037). Differences in DNA concentration were detected depending on the appearance of cytological samples, with higher values for bloody samples, followed by slightly bloody, and finally, turbid-clear appearance (P<.001), according to the cell contents.
Lymph Node Characteristics Based on the Concentration of DNA Initially Extracted and After Treatment With Sodium Bisulfite.
| Variable | Category | DNA (ng/μL) median and IQR | Treated DNA (ng/μL) median and IQR |
| Gender | Male (n=147) | 43.50 (25.70–102.20) | 19.90 (8.70–60.70) |
| Female (n=23) | 22.50 (7.90–74.20) | 9.90 (3.70–59.00) | |
| P=.037a | P=.196a | ||
| Appearance of punctured sample | Clear-turbid (n=76) | 27.35 (14.98–64.10) | 10.90 (6.05–30.70) |
| Slightly bloody (n=65) | 42.80 (30.20–84.70) | 18.60 (5.15–54.15) | |
| Bloody (n=29) | 157.40 (50.65–356.05) | 82.60 (33.85–142.95) | |
| P<.001b | P<.001c | ||
| DNA purity (ratio 260nm/280nm) | 1.7–2.0 (n=104) | 61.50 (32.23–174.88) | 36.95 (17.30–100.48) |
| >2.0 (n=14) | 7.10 (5.00–9.65) | 3.85 (1.48–5.68) | |
| <1.7 (n=52) | 36.85 (23.80–50.78) | 9.65 (4.15–18.46) | |
| P<.001d | P<.001d | ||
| Purity of bisulfite-treated DNA (ratio 260nm/280nm) | 1.8–2.2 (n=108) | 64.20 (36.85–164.95) | 35.30 (16.01–96.85) |
| >2.2 (n=45) | 25.60 (12.25–41.80) | 9.2 (3.6–14.20) | |
| <1.8 (n=17) | 22.20 (9.10–37.95) | 4.50 (1.90–14.80) | |
| P<.001e | P<.001f | ||
| Node station punctured | Mediastinal (n=147) | 42.70 (22.80–102.20) | 19.90 (7.70–60.90) |
| Hilar (n=23) | 42.80 (16.80–64.20) | 10.80 (7.00–37.60) | |
| P=.483a | P=.512a | ||
| Node station punctured | 4R+7 (n=124) | 41.50 (22.58–98.50) | 18.60 (8.55–60.85) |
| 4L. 11L. 11R. 8. 2R. 10R y 12R (n=46) | 45.65 (19.48–101.93) | 20.10 (6.98–50.45) | |
| P=.851a | P=.833a | ||
| Node station punctured | 4R (n=67) | 41.10 (24.50–74.20) | 17.90 (7.70–45.60) |
| 7 (n=57) | 43.50 (22.45–109.80) | 19.60 (8.60–74.60) | |
| 4L (n=16) | 30.00 (17.03–104.20) | 19.10 (3.78–66.68) | |
| 11L (n=12) | 36.60 (13.65–64.20) | 9.10 (6.93–29.73) | |
| 11R (n=8) | 45.65 (19.15–50.08) | 25.83 (4.88–36.93) | |
| 8 (n=4) | 103.55 (74.03–282.03) | 47.35 (40.20–141.50) | |
| 2R (n=3) | 50.30 (40.10–101.40) | 36.90 (11.50–37.00) | |
| 10R (n=2) | 104.90 (42.80–167.00) | 79.85 (42.20–110.50) | |
| 12R (n=1) | 11.30 | 10.80 | |
| P=.439g | P=.369h | ||
| Nodal diagnosis | Benign (n=82) | 41.00 (16.40–84.15) | 17.90 (4.75–41.98) |
| Adenocarcinoma (n=54) | 40.90 (16.80–78.40) | 17.90 (4.80–40.50) | |
| Epidermoid (n=28) | 46.55 (30.10–221.45) | 24.25 (12.33–93.93) | |
| Large cell (n=6) | 138.45 (33.30–363.33) | 65.85 (12.33–198.68) | |
| P=.163i | P=.164i | ||
| Metastases | Yes (n=88) | 42.95 (24.78–137.35) | 22.00 (7.00–58.55) |
| No (n=82) | 41.95 (19.18–88.38) | 17.41 (7.68–60.75) | |
| P=.390a | P=.093a | ||
IQR: interquartile range.
The table shows the P-values obtained with the Kruskal–Wallis test. Post-hoc to the Kruskal–Wallis test, Mann–Whitney U was performed, with a paired comparison of the categories for each variable.
The DNA extraction protocol used provided optimal or good purity (ratio 260/280nm) in 61.18% and 8.24% of cases, respectively. Higher DNA concentrations were obtained from optimal purity samples, followed by those with contaminants, and finally, good purity samples (P<.001, Table 1).
Regarding the node station punctured, no differences in DNA concentration were found when comparing hilar and mediastinal lymph nodes (P=.483), nor when lymph nodes mostly from stations 4R and 7 were compared with the rest (P=.851). No differences in the concentration of DNA were observed between lymphadenopathies according to site. However, lymphadenopathies from station 8 had a higher DNA concentration compared to stations 4R, 11R and 11L (Table 1). DNA concentration did not have normal distribution or homogeneity of variance over the diagnostic groups, with medians of approximately 40ng/μL for the benign, adenocarcinoma and squamous cell categories, and 138.4ng/μL for the six cases of large cell tumor (Table 1). Consequently, no differences were detected in DNA concentration in terms of nodal diagnosis (P=.163) or between benign and malignant lymph nodes (P=.390).
Characteristics of Deoxyribonucleic Acid After Sodium Bisulfite TreatmentThe concentration of modified DNA in relation to some variables was similarly studied (Table 1). The concentrations recorded were lower than those observed before treatment, ranging from 0.20 to 392.30ng/μL (median: 19.13ng/μL, IQR: 7.60–60.18ng/μL). Comparing the concentrations before and after treatment, significant differences (P<.001) were observed, with lower concentrations in 93.53% of the samples.
Although men continued to show higher DNA concentrations than women, the differences were not significant (P=.196). Concentrations of modified DNA differed significantly, depending on the appearance of the samples (P<.001). Moreover, the purity estimation of the modified DNA indicated that 63.53% and 26.47% were of optimal and good quality, respectively, with up to 90% of the samples potentially suitable for methylation analysis. The median for optimal purity samples was the highest, at least three times higher than that of the samples of lower purity (P<.001).
Regarding the lymph node station punctured, no difference was detected between the mediastinal and hilar lymphadenopathies (P=.512), nor between stations 7 and 4R compared to the rest (P=.833), as shown in Table 1. Similarly to DNA concentration before treatment, the differences in modified DNA were found in lymphadenopathies 2R, 4R, 11R and 11L compared to those from station 8 (Table 1). Finally, no differences were found when comparing the nodal diagnostic categories (P=.164) or benign and malignant lymph nodes (P=.093) (Table 1).
Methylation Studies From Lymph Node Cytology SamplesAlthough most samples seemed adequate for further studies, validity was confirmed with methylation analysis of two tumor suppressor genes, using two different methodologies. Table 2 summarizes sample characteristics according to the methylation data collected (samples with/without methylation data) for both genes and the two approaches.
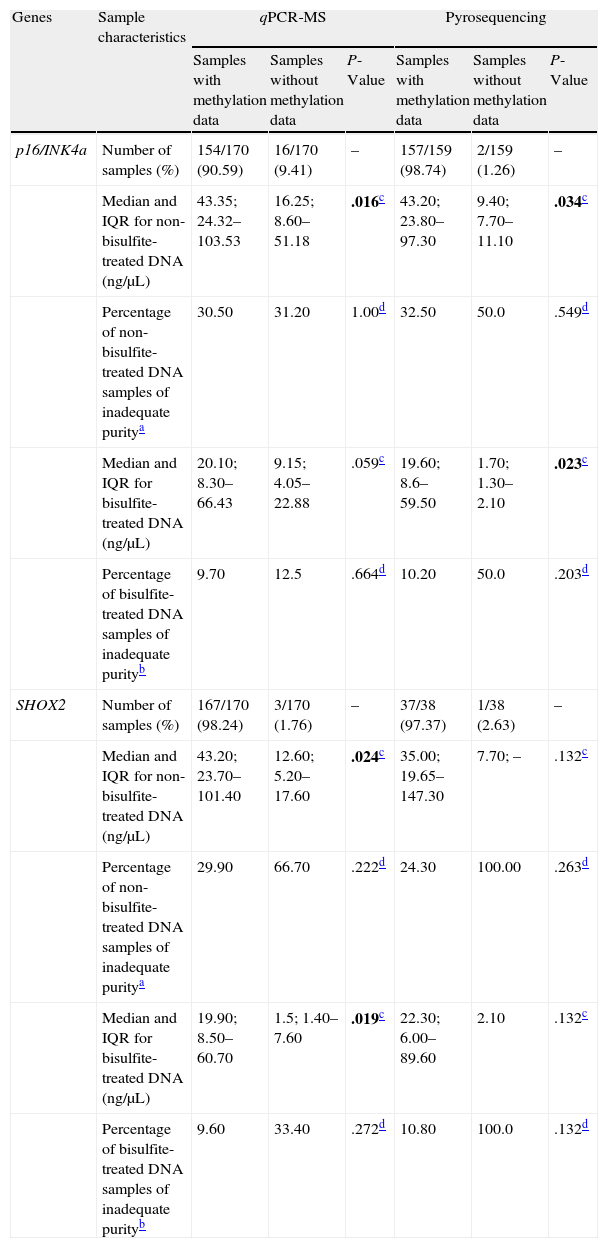
Characteristics of Lymph Node Samples for p16/INK4a and SHOX2 Methylation Analyses.
| Genes | Sample characteristics | qPCR-MS | Pyrosequencing | ||||
| Samples with methylation data | Samples without methylation data | P-Value | Samples with methylation data | Samples without methylation data | P-Value | ||
| p16/INK4a | Number of samples (%) | 154/170 (90.59) | 16/170 (9.41) | – | 157/159 (98.74) | 2/159 (1.26) | – |
| Median and IQR for non-bisulfite-treated DNA (ng/μL) | 43.35; 24.32–103.53 | 16.25; 8.60–51.18 | .016c | 43.20; 23.80–97.30 | 9.40; 7.70–11.10 | .034c | |
| Percentage of non-bisulfite-treated DNA samples of inadequate puritya | 30.50 | 31.20 | 1.00d | 32.50 | 50.0 | .549d | |
| Median and IQR for bisulfite-treated DNA (ng/μL) | 20.10; 8.30–66.43 | 9.15; 4.05–22.88 | .059c | 19.60; 8.6–59.50 | 1.70; 1.30–2.10 | .023c | |
| Percentage of bisulfite-treated DNA samples of inadequate purityb | 9.70 | 12.5 | .664d | 10.20 | 50.0 | .203d | |
| SHOX2 | Number of samples (%) | 167/170 (98.24) | 3/170 (1.76) | – | 37/38 (97.37) | 1/38 (2.63) | – |
| Median and IQR for non-bisulfite-treated DNA (ng/μL) | 43.20; 23.70–101.40 | 12.60; 5.20–17.60 | .024c | 35.00; 19.65–147.30 | 7.70; – | .132c | |
| Percentage of non-bisulfite-treated DNA samples of inadequate puritya | 29.90 | 66.70 | .222d | 24.30 | 100.00 | .263d | |
| Median and IQR for bisulfite-treated DNA (ng/μL) | 19.90; 8.50–60.70 | 1.5; 1.40–7.60 | .019c | 22.30; 6.00–89.60 | 2.10 | .132c | |
| Percentage of bisulfite-treated DNA samples of inadequate purityb | 9.60 | 33.40 | .272d | 10.80 | 100.0 | .132d | |
IQR: interquartile range. Significant P-values highlighted in bold.
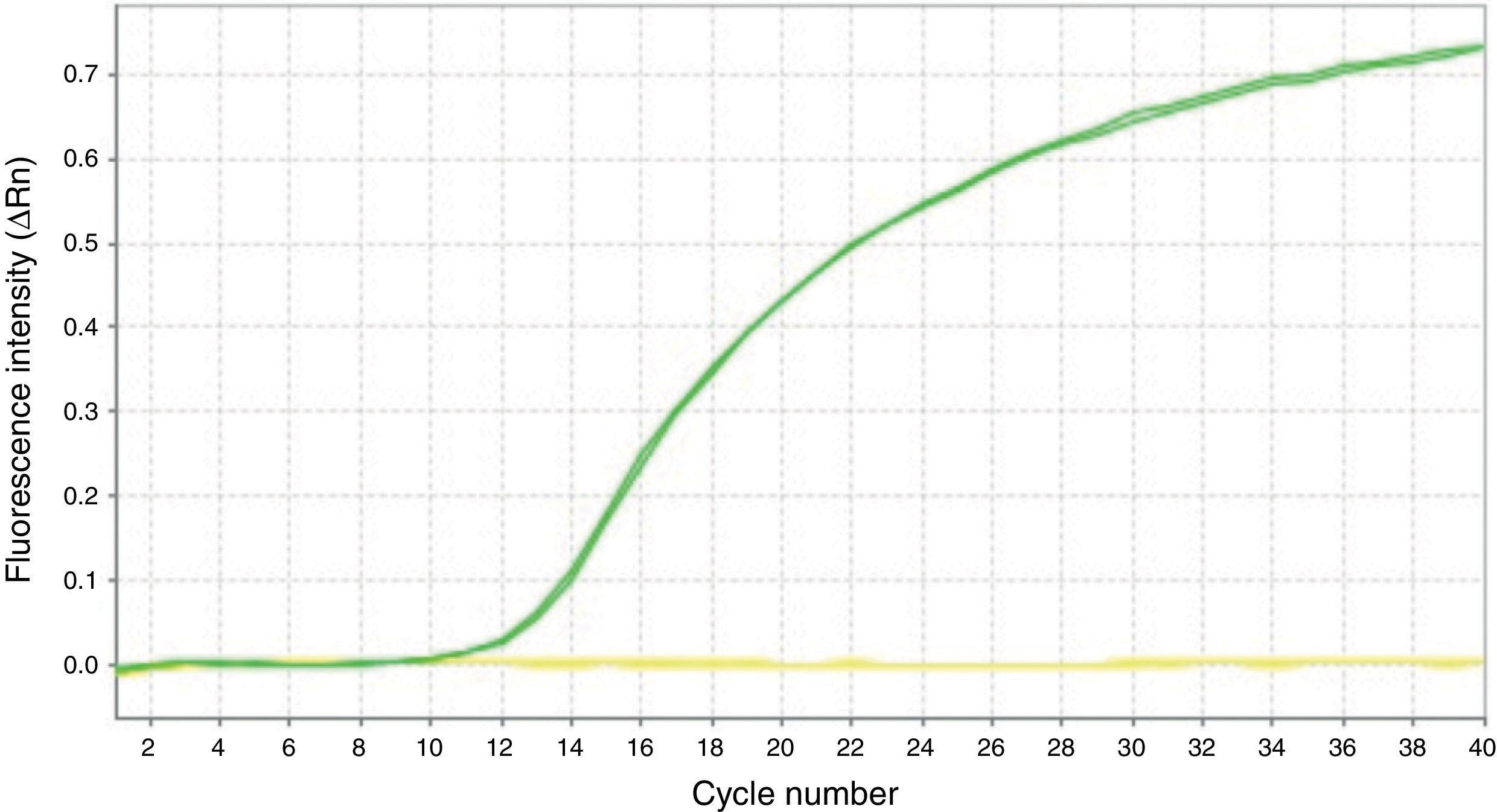
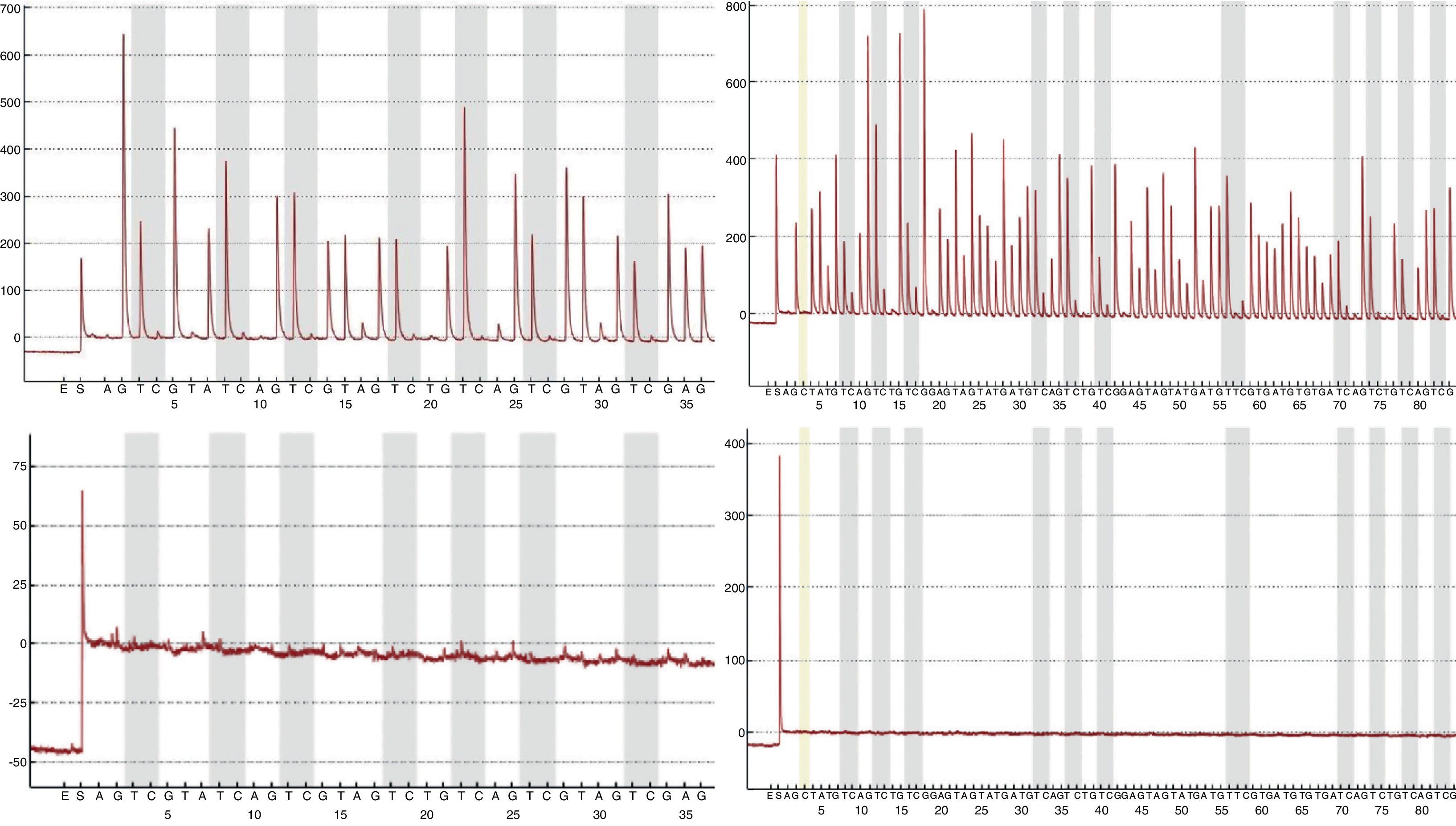
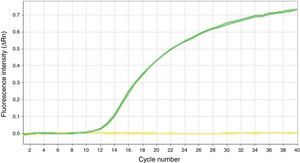
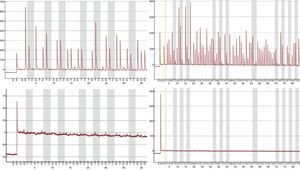
The MS-qPCR technique did not allow analysis of 9.41% (16/170; p16/INK4a) and 1.76% (3/170; SHOX2) samples, whereas pyrosequencing provided no information in 1.26% (2/159; p16/INK4a) and 2.63% (1/38; SHOX2) of cases. DNA concentration medians and IQRs before and after treatment were significantly lower in the samples without methylation data compared to those in which these data were available. This difference was statistically significant (Table 2) and related to the difficulty in testing certain samples. However, the percentage of samples with inadequate purity varied widely both for samples without methylation data and for those with data; this was greater in the first group in all cases and was also associated with the impossibility of obtaining results with the techniques used. Fig. 1 shows the qPCR amplification curves for p16/INK4a in a valid and an invalid sample. The pyrograms of p16/INK4a for a valid and an invalid sample are shown in Fig. 2.
p16/INK4a pyrograms of a valid sample (top panels) and an invalid sample (bottom panels). Pyrosequencing was performed in two different reactions to cover the 18 CpG sites of the island: pyrograms on the left show the first seven CpG sites, and on the right, the last eleven CpG sites. The gray bars in the pyrograms indicate each CpG site.
There is ample evidence of occult micrometastases in lymph nodes that are undetectable by standard cytohistologic methods.5,9–12 The recent introduction of molecular techniques has led to an improvement in the detection of metastatic cells, in both tissues, including cytological material, and various biological fluids.9 Recently, Nakajima et al.11 reported that DNA, RNA and protein can be simultaneously isolated from samples obtained by EBUS. However, at present few published data are available on the molecular analysis of these samples,5,10,11 including expression studies by qPCR of cytokeratins, MUC1, p53, K-ras, CEA, CK19, KS1/4, lunx, PDEF and ALK13–15; mutation detection in EGFR, KRAS, BRAF and PIK3CA16; and cytokeratins and ALK detection by immunohistochemistry.13,15,17 Also, given the importance of aberrant methylation of CpG islands as the main epigenetic mechanism for the regulation of gene silencing, and its involvement in neoplastic processes, molecular studies have also focused on this area, studying the methylation status of the MGMT, p16/INK4a, p14/ARF, FANCF, Reprimo, TMS1/ASC and AP-2α genes.9,18
It can be seen from this paper that lymph node samples obtained by EBUS are suitable for methylation studies using MS-qPCR and pyrosequencing techniques, and that greater sensitivity is obtained than with the MS-PCR commonly used in methylation analysis, including the above-referenced studies.9,18
Unlike other molecular studies of blood samples, the initial disadvantage of using samples obtained by EBUS is the limited volume recovered in each puncture, along with the variations in cellularity, so extraction of as much good quality DNA as possible is a priority. The protocol used in our study allowed the retrieval of optimal/good purity DNA in 69% of samples. In addition, no changes were observed for the node station punctured or the final histological diagnosis of the malignancy.
Unlike mutations accompanied by DNA sequence changes, DNA methylation consists of the transfer of methyl groups (CH3) to DNA cytosine bases located before and next to a guanine. Therefore, a prerequisite for the study of methylation is treatment with sodium bisulfite, which converts all unmethylated cytosines to uracil, whereas methylated cytosines remain unaltered.19,20 This modification is essential to distinguish methylated cytosines from the unmethylated form.
During bisulfite treatment, DNA is subjected to aggressive conditions (long incubations at high temperatures and acidic pH, high molarities of bisulfite and urea) to ensure complete conversion,20 which is a critical point for correct methylation analysis, as overestimation is avoided.21 The kit used for bisulfite treatment in this study is designed to minimize DNA degradation and loss. The optimal amount of starting DNA is 200–500ng (10–25ng/μL); only 8.82% of the study samples had lower amounts, although these were still within the recommended wide range (50pg–2μg).
Although the estimated initial DNA recovery with this kit is more than 80%, 93.53% of the treated samples produced lower concentrations, with a mean decrease of 52.92±38.77%. Despite this reduction, the quality of the modified DNA was optimal/good in 90% of the samples. Ehrich et al. (2007),20 using the same bisulfite kit used in this study, designed an assay to determine the quality of modified DNA, and found that good amplification was achieved in fragments up to 477bp in size, with reduced quality in those sized 617bp.
While the quantity and quality of modified DNA are apparently appropriate for further study, sample validity was verified using MS-qPCR and pyrosequencing. MS-qPCR is a quantitative method that reports the percentage of methylation on the entire CpG island, while pyrosequencing is used as gold standard in the methylation analysis, and provides the percentage of methylation for each site studied in the CpG island.
The selected genes were p16/INK4a and SHOX2, widely described in the literature as potential methylation biomarkers for prognosis and diagnosis of LC.22–25p16/INK4a is involved in cell cycle regulation, inducing its interruption, while SHOX2 belongs to the homeobox gene family and is related to DNA packing. Methylation of these genes has been studied with MS-PCR, MS-qPCR, pyrosequencing and restriction enzymes sensitive to methylation sites, among others, using primary tumor samples, serum, plasma, sputum, aspirate, or bronchial lavage samples.22,23,25
The CpG island studied in p16/INK4a has 150bp and includes 18 CpG sites, whereas that of SHOX2 has 124bp and 11 CpG sites, so they are close to the 150–300bp range recommended for good quality amplification.20,21 Quantification of methylation was not possible in some samples due to the low DNA yield. This, added to inadequate purity, fragmentation and degradation as a result of treatment, explains the difficulty in performing the analysis.
A DNA concentration 2.6 and 4.6 times higher in samples with methylation data compared to that in samples without data highlights the importance of the initial amount of DNA. The differences were even greater for DNA concentration after bisulfite treatment, between 2.2 and 13 times higher. According to our study, a DNA concentration from EBUS samples of at least 20ng/μL is desirable for sodium bisulfite treatment. Also, in order to be able to quantify methylation by nested MS-qPCR or pyrosequencing, a concentration of treated DNA not less than 8ng/μL is advisable.
It is reasonable to mention that, in MS-qPCR analysis, the absence of p16/INK4a methylation probably explains the greater number of samples without data, since analysis by pyrosequencing was possible in almost all samples. For SHOX2, although pyrosequencing was not possible in 2.63% of the samples, only 38 samples were pyrosequenced, while only one sample could not be pyrosequenced, either for SHOX2 or for p16/INK4a.
Advances in less invasive sampling techniques such as EBUS, along with a better understanding of the molecular basis of tumor development and progression, such as aberrant DNA methylation, are tools that together represent a promising strategy for improvement in the diagnosis and staging of LC. Translation of this strategy into clinical practice requires suitable starting material in terms of DNA quantity and quality. According to this study, a multidisciplinary approach is possible that would result in a high level of retrieval, as MS-qPCR analysis (nested PCR) could be performed on up to 12 methylation markers. Moreover, if sufficient good quality material is obtained, a combined marker panel could be designed that would definitely improve the sensitivity required for the detection of microscopic lymph node involvement (occult N) that is unachievable with current cytohistological techniques.
ConclusionQuantification of methylation by techniques such as MS-qPCR and pyrosequencing from lymph node samples obtained by EBUS-TBNA is feasible, provided that minimally adequate DNA concentrations can be obtained. The possibility of carrying out epigenetic studies in lymph node samples obtained by EBUS might significantly contribute to the search for epigenetic biomarkers that will improve decision making in potentially curable LC, to the patients’ benefit.
FundingSector Program for Applied Research (INCITE, Consellería de Innovación e Industria de la Xunta de Galicia). Code 09CSA053905PR.
Health Research Funds of Instituto Carlos III. Code PI09/90385.
The samples used belong to CHUVI biobank (RETIC-FIS-ISCIII RD09/0076/00011).
E.U. Seventh Framework Program (FP7/REGPOT-2012-2013-1. Code: N.o 316265, BIOCAPS.
AuthorshipStudy conception and design: Leiro-Fernández, De Chiara, Fernández-Villar, Botana-Rial.
Patient recruitment: Leiro-Fernández, Núñez-Delgado, Botana-Rial, Fernández-Villar.
EBUS-TBNA performance: Leiro-Fernández, Fernández-Villar, Núñez-Delgado, Botana-Rial.
Pathology analysis: Tardío-Baiges, González-Piñeiro.
Molecular determinations: De Chiara, Valverde-Pérez.
Data analysis and interpretation: Leiro-Fernández, De Chiara, Fernández-Villar, Leiro-Fernández, González-Piñeiro, Tardío-Baiges.
Statistical analysis: De Chiara, Leiro-Fernández, Fernández-Villar.
Manuscript writing: De Chiara, Leiro-Fernández, Fernández-Villar.
Critical review of manuscript: Valverde-Pérez, Núñez-Delgado, Botana-Rial.
Study supervisor: Fernández-Villar.
Conflicts of InterestThe authors declare no conflicts of interest.
Please cite this article as: Leiro-Fernández V, De Chiara L, Botana-Rial M, González-Piñeiro A, Tardio-Baiges A, Núñez-Delgado M, et al. Viabilidad de las muestras ganglionares obtenidas por ecobroncoscopia para el estudio de alteraciones epigenéticas en pacientes con cáncer de pulmón. Arch Bronconeumol. 2014;50:213–220.