It is increasingly common to have medical information in electronic format. This includes scientific articles as well as clinical management reviews, and even records from health institutions with patient data. However, traditional instruments, both individual and institutional, are of little use for selecting the most appropriate information in each case, either in the clinical or research field. The so-called text or data “mining” enables this huge amount of information to be managed, extracting it from various sources using processing systems (filtration and curation), integrating it and permitting the generation of new knowledge. This review aims to provide an overview of text and data mining, and of the potential usefulness of this bioinformatic technique in the exercise of care in respiratory medicine and in research in the same field.
Cada vez es más habitual disponer de información médica en formato electrónico. Esto incluye tanto artículos científicos como revisiones sobre el manejo clínico e incluso registros de instituciones sanitarias con datos de pacientes. Sin embargo, los instrumentos tradicionales, tanto individuales como institucionales, son poco útiles para seleccionar la información más apropiada en cada caso, sea en el ámbito clínico o en el de la investigación. La llamada «minería» de textos o de datos permite gestionar esa gran cantidad de información, extrayéndola de fuentes diversas mediante sistemas de procesamiento (filtrado y curado), integrándola y permitiendo la generación de nuevo conocimiento. La presente revisión pretende proporcionar una idea general sobre la minería de textos y datos, así como sobre la ayuda que esta técnica bioinformática puede suponer para el ejercicio asistencial de la medicina respiratoria y para la investigación en ese mismo campo.
The high financial and social burden of the most common respiratory diseases [bronchial asthma, chronic obstructive lung disease (COPD), infections and lung cancer] represents a great challenge to health and healthcare services.1–7 Advances in information technology mean that enormous amounts of health-related data can now be accessed and analyzed. It is common for patient notes to be recorded in electronic format in both hospitals and primary care centers. This includes not only personal data, but also diagnoses, severity, laboratory test results, function tests and medication, and details on the patient's contacts with the healthcare system. There are three important advantages in recording this large amount of data in digital format: (i) quality is improved, (ii) the time healthcare workers spend on unproductive tasks is reduced, and (iii) the data can be used in automatic systems, such as “text mining” or “data mining”.8,9 Strictly speaking, the difference between text and data mining is that in the first, information is obtained from free text formats, while in the second, it is obtained from databases. One example of the pioneer use of clinical data on an institutional level is the MedLEE platform,10 an automatic system that allowed relevant information to be drawn from clinical case reports.
Other highly important areas that can benefit from computer-based tools are professional training and research. It is difficult for the medical professional in these fields to manage and sort the vast amount of information available from numerous sources. Indeed, numerous publications in both the classic paper format and the electronic format have attempted to recompile the most significant events in the publishing world on specific areas of medical knowledge. The UpToDate series,11 for example, has published some issues dedicated to respiratory medicine (Pulmonary, Critical Care and Sleep Medicine, and Allergy and Immunology, respectively). The electronic mail alert systems aimed at professionals with certain user profiles work in a similar way. Text mining instruments are also used in these cases, allowing greater depth in the search, selection and processing of data.
Systems Biology and MedicineSystems biology has been defined as the science of the systematic study of interactions in biological processes. When talking specifically of human diseases, the expression used is Systems Medicine, although this term also has other meanings. For advances to be made in Systems Biology and Medicine, connections must be made between the bodies of knowledge available in the different scientific fields.12 This way, new properties and hypotheses that could not be identified by the traditional approaches may come to light. The instruments that facilitate this interdisciplinary task are to a large extent the product of the information technologies that enable the processing of large amounts of diverse data, thereby generating new knowledge and information. Systems Biology and Medicine methods can be used to establish mathematical models of disease that integrate structural and physiopathological knowledge in their different levels of complexity.13 The aim of this approach is not only to achieve a greater understanding of nosological processes, but also to possibly simulate body systems for generating new diagnostic and therapeutic approaches.
Various instruments are used in Systems Biology and Medicine. One of these, and the main focus of this review, is text mining. Text mining can be defined as a set of information technologies for detecting, extracting and interpreting, automatically (or semiautomatically), digital information that is basically in text format. In addition, there are the above-mentioned mathematical models of biological or nosological processes. To produce these models, instruments and procedures must be used that can adequately process the large amounts of data obtained both from advanced biological analytical techniques and from large clinical and/or epidemiological studies. Among the biomedical disciplines and techniques associated with Systems Biology and Medicine are genomics (study of the genes), transcriptomics (study of transcriptomes), proteomics (structure and function of proteins), interactomics (molecular interactions and consequences), metabolomics (molecular signals left by biological processes) and metagenomics (sets of genomes occurring together in an environment); in other words, the disciplines popularly known as the “omics”. In the area of medicine and epidemiology, there are several examples of these techniques being used in relation with the respiratory tract. Some of the most recent were the papers published by Garcia-Aymerich et al.14 and Burgel et al.15 on COPD phenotyping, obtained from a cluster analysis of all kinds of data obtained from real patients. Yet another facet of Systems Biology and Medicine, also related with the “omics”, is the analysis of results generated by techniques such as the microarrays or chips used for the simultaneous and respective analysis of the differential expression of large series of DNA and protein. These techniques have already been widely used in respiratory disease research. Thus, Steiling et al.16 and Pierrou et al.17 demonstrated the effects of smoking on the expression of multiple genes associated with cell damage and oxidative stress in the bronchial epithelium. Some studies, refining the complexity even further, combine genetic data with clinical and epidemiological data.18
Text MiningAs already mentioned, text or data mining constitutes a set of computerized techniques that allow the automatic processing of digital information. Using these systems, information is processed into a manageable format, and new knowledge is also generated. This instrument has been used for years in other biomedical fields, such as molecular biology or systems biology itself, where it has contributed interesting results.9,19 Text mining, in its most simple form, is used by anyone who employs tools such as PubMed20 on a literature database such as MEDLINE, selecting an area of interest from keywords or certain authors.21
After the relevant information has been obtained using text mining techniques, the next step is “curation”. This phase can be carried out in an automatic, semi-automatic or manual fashion. In automatic curation, additional requirements can be added, whether binary (information is selected or rejected according to pre-established rules) or categorical [for example, specific weight is given to the impact factor of a journal, the type of article, the design or level of evidence of the study or the origin of the data (humans, animal models, cell lines), etc.]. In manual curation, an expert in the subject matter cleans the data lists or information sources, according to his/her criteria. However, as discussed below, all too often any biomedical scientist is considered an expert, even if they are not a specialist in the area under investigation.
A third important point is that once the information has been selected, connections must be established between diverse data. This process is called integration and is essential for generating new knowledge. It is obvious that the connection and integration of data obtained from different areas of expertise do not often emerge spontaneously using the traditional methods. Accordingly, automatic methods that select and put potentially significant information at the disposal of the professional are necessary, regardless of the area of knowledge in which it has been generated.
In the sections below, more details on how text or data mining can be used in the different areas of medicine will be discussed, including how significant findings in the more basic research can be translated to the clinical management, diagnostics and severity classification, and prognosis of some of the most common respiratory diseases.
Historical ContextualizationAlthough the need for the automatic processing of large amounts of information is relatively new, the technology on which the current text mining techniques are based has been around for some time. Indeed, half a century ago, similar techniques were already being used for discourse analysis22 and linguistic structure studies.23 In medicine, some of the pioneer works were carried out by Swanson,24,25 who established relationships between apparently unconnected phenomena, such as migraine and magnesium deficiency, from the titles of articles obtained from the MEDLINE data base.25 These connections were validated years later in experimental studies.26 Using similar methodology, Srinivasan and Libbus27 and Weeber et al.28 uncovered the potential beneficial effect of curcumin and thalidomide in patients with Crohn's disease. These types of search are now widely used in medicine and in biology9 for establishing associations between, for example, genes (or proteins) and diseases,29–32 or for evaluating interactions between various proteins.33,34 An interesting instrument for this purpose is DisGeNET, designed to relate information derived from the various “omics” with data generated on different diseases.31,32 As for respiratory diseases, as discussed below, these techniques and instruments are being used intensively in the study of various aspects of COPD, bronchial asthma and lung cancer.
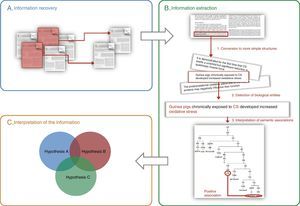
General Aspects of Text MiningThe purpose of text and data mining is, by definition, to recover, extract and interpret information stored in electronic format in large documentation archives and databases, using automatic or semiautomatic tools (Fig. 1).9
General schematic of the methods used in text and data mining: (A) information recovery, (B) information extraction, (C) interpretation of the information (the integration of various previously corroborated hypotheses can produce a new combined hypothesis). The three steps required for extracting information are shown in panel B: (1) breakdown of the information into basic units (e.g. sentences), (2) identification of biological entities, and (3) interpretation of the relationships between biological entities.
Information recovery consists of identifying documents that contain data on the question of interest; for example: “Which biomarkers have been involved or are potentially involved in the diagnosis of COPD?” Currently, locating the information is not a problem, since there is an endless source of data accessible in electronic format, whether original (journals, text books, websites) or already recompiled in databases. One example of a database source is MEDLINE, accessible through the PubMed information recovery system.20 Some more specific systems are available, such as Textpresso35,36 or HLungDB,37,38 a lung cancer database that pools information on implicated genes and proteins.
The aim of the second step, information extraction, consists of identifying the relevant part of the information from all the recovered data. In essence, three stages are required (Fig. 1B): (1) processing, with the conversion of complex texts into simple structures (e.g. words or short sentences) that can be interpreted by the computer systems, (2) firm identification (standardized) of the clinical entities or biological processes referred to in the documents, and (3) interpretation of the semantic relationships between diverse structures or entities. For extracting information, systems based on artificial intelligence algorithms,39 statistical methods40 or mixed systems (e.g. GENIA Tagger41,42 or Standford Log-Linear Part-of-Speech Tagger43,44) are generally used.
A problem inherent to the identification and interpretation phases is that the automatic mechanisms that allow filtration of the available knowledge do not have enough discretional criteria. Add to this the fact that, in general, the teams of professionals who design these instruments lack experts in the specific search subjects. The gap between the latter and the bioinformatics specialists is even greater in the case of medicine than for biology, and has been identified as the principal problem in the design of filtration and curation instruments. This is a problem inherent to the current pattern of fragmenting science into disciplines, and only becomes more accentuated as knowledge becomes progressively more segmented within each specialty. Thus, it is essential to include clinical professionals in multidisciplinary working teams.45
An additional problem is that the results of the studies are not axioms, but are frequently treated as such by professionals trained in more exact sciences than medicine. It is also important to remember that although text mining is not a totally new technology, and its results are promising, there is still room for improvement. For example, on a technological level, there is a problem inherent to biomedical language that is the lexical and semantic variability among the different disciplines. Although this problem can be approached with some success with artificial intelligence algorithms or statistical methods, it has not been fully resolved. Another aspect to take into consideration, primarily in the use of text mining in medicine, is that the conclusions can end up having consequences for patients, so it is important that they are fully corroborated.
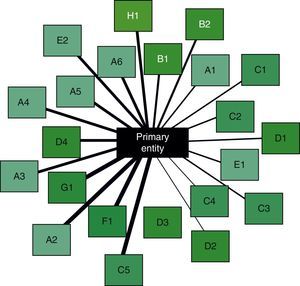
The final step is the interpretation of results. This is aimed at integrating the information obtained in the previous steps, ultimately to obtain associations between disease entities or biological or clinical phenomena that were initially undetectable using conventional methods. Returning to the Swanson study,25 the integration of individually verified hypotheses brought to light a new hypothesis, linking magnesium deficiency with migraine. By way of example, Fig. 2 is a schematic of the associations between an entity (COPD) and its comorbidities. These types of images help in the simultaneous interpretation of diverse information obtained with data mining, leading to the generation of new hypotheses.
Schematic of the relationships between a primary entity (center) and several potential comorbidities (boxes around the edge). The thickness of the lines represents the amount of information available in each case, after automatic curation of the selected data. Their significance has been arranged in a clockwise order of relevance. Comorbidities in the same body system are shown with the same color in the background of the box and with a corresponding identification letter.
As mentioned above, the high production and availability of data in the area of medical and more basic biological sciences make it an appropriate space for the use of text mining. In the specific field of respiratory diseases, numerous studies making use of this technique have appeared in recent years, covering the various stages from the most basic knowledge to applied medicine.
- Knowledge in basic sciencesIn fields such as molecular biology or physiology, text mining is bringing about a transition from the classic deductive scientific model, in which a hypothesis is tested experimentally, to a model based on the “blind” search for associations between apparently unconnected facts that can later be experimentally validated.46,47 For this approach, the development of specialized platforms is desirable. Thus, we have combined databases, such as the above-mentioned HLungDB,37,38 that have integrated information on genes, proteins, epigenetic modifications and clinical characteristics, related in all cases with lung cancer. The main aim of this platform is to establish a network of connections for molecules associated with this disease entity, thus facilitating not only more basic research but also integration with relatively distant areas, such as clinical medicine or epidemiology. This is essential for diseases such as lung cancer that involve complex alterations and multifactorial causes. In this way, previously unexplored research pathways can be developed and new therapeutic alternatives can be suggested. There are also numerous examples of the use of text mining in the basic aspects of COPD. Comandini et al.48 used data on genetic and protein expression in non-smokers, smokers without lung disease and smokers with COPD, to identify tobacco-induced effects. Their results suggest that certain genes with antioxidant activity are over-expressed in the presence of tobacco smoke in some individuals, and this may play a significant role in protecting tissue from oxidative stress. These genes or the products of these genes may be used as negative biomarkers for the risk of developing COPD. In a very recent study by our group,49 text mining was used to study the comorbidities of this same entity and the mechanisms that might be involved. This threw up a surprising amount of data for some of the associations (such as COPD with ischemic heart disease or lung cancer) and a scarcity of data in the literature for others (as is the case for COPD and nutritional changes or pulmonary hypertension). Some interesting work has also been published in the area of bronchial asthma. For example, Su et al.50 investigated how various genes interrelated among themselves and with the environment in childhood asthma, and concluded that some genes determine the susceptibility of the carrier for developing childhood asthma. In another study, Tremblay et al.51 developed a methodology (Genes to Diseases, G2D) based on text mining for identifying genes that were possibly involved in the development of asthma and atopy. Something similar is occurring with pneumonia and adult respiratory distress syndrome (ARDS). By analyzing multiple markers in bronchoalveolar lavage samples of ARDS patients, Frenzel et al.52 showed that elevated interleukin-6 had great prognostic value.
It is clear then that the automatic analysis of knowledge from various basic sciences can shed new light on respiratory diseases.
- Diagnosis and clinical management of the respiratory patientIt is crucial in any disease to establish a definitive diagnosis and to classify severity. However, this is not always an easy task, particularly if the diagnosis requires the deployment of complex techniques or if special training is required. This is the case in COPD, where patient collaboration and a satisfactory forced spirometry procedure are essential. At the present time, according to some initiatives such as the Global Initiative for Chronic Obstructive Lung Disease (GOLD),53 the number of recent hospital admissions and the grade of dyspnea must also be determined. The adequacy of a certain spirometric maneuver can be verified by automatic data analysis, while a complete diagnosis based on other variables is obtained. Matsumoto et al.54 used data mining on the electronic records of some 27,000 patients to show that airway obstruction detectable by spirometric values had not been diagnosed in up to 86% of cases. These are some examples of the application of data mining that, moreover, allow diagnostic alerts to be flagged in clinical charts, preventing significant findings from going unnoticed by health professionals. Furthermore, as we have seen, the automatic analysis of data allows new associations between variables to be established, generating hypotheses that can lead to alternative or complementary diagnostic systems. With regard to lung cancer, there are several critical steps in the detection and subsequent stratification of this disease. One of these is the evaluation of lymph node involvement that is initially carried out using imaging techniques, namely, computed axial tomography (CAT) and positron emission tomography (PET) scans, and confirmed or ruled out from tissue samples obtained by endobronchial ultrasound or mediastinoscopy. Lu et al.55 proposed a new automatic method for the initial orientation of lymph node involvement. This is also based on CAT data, but data mining is then used to compare the findings with the anatomical registries of lymph nodes from already diagnosed patients.
One phase in the diagnostic process that is complementary to the identification of the disease is establishing prognosis. This generally involves categorizing disease severity or extent. It is equally important at this stage in the process to have good classification methods, since this will frequently determine the treatment offered. Automatic lung cancer staging systems that use clinical and histological data recorded in diverse reports from one or several institutions have been developed in recent years.56,57 For lung or combined heart–lung transplantation, an accurate prediction of survival is essential, not only for a specific patient, but also for the selection of donors and recipients. A vast amount of information is currently available on transplantations (procedures, patient monitoring, etc.) and this has been used along with classic statistical methods to make morbidity and survival predictions for different organs, including lungs.58 More recently, data mining has also been used,59 since it has several advantages over the classic statistical methods. For example, it is not limited by the number of observations, nor does it need observer independence, as would occur in a specific study.
One area in which data mining has been particularly fertile is in the care of semicritical and critical respiratory patients. Over the last decades, the units caring for these patients have set up data collection systems, and as a direct consequence, two major applications based on text mining have been developed. One is the generation of short- and mid-term predictive models that allow the creation of intelligent alerts and decision-making systems. Among the numerous studies in critical patients, of particular importance is that of Tzavaras et al.,60 who used data mining and neural networks to develop a clinical decision support model, based on the identification of key physiological variables for determining when mechanical ventilation should be initiated.
Another application is the development of long-term models, mainly used as methods for evaluating the quality of the teams and institutions involved in the care of the critical respiratory patient.61–63 Some of these applications have been subsequently extended to conventional hospital units, such as pulmonology and thoracic surgery wards. Traditionally, these instruments were constructed using only demographic and administrative data to determine survival rates, mortality, etc. However, more recently, archived physiological and clinical data have been used to generate data mining-based models. There are several studies, such as those of Bohensky et al.62 or Kim et al.63 that show that these models are superior to the classical methods in the prediction of survival.
Clinical pathways are the records of the procedures performed on specific patients during their hospital stay. Studies using automatic analysis methods have also been published in this field.64,65 Clinical pathways are a basic tool in healthcare quality management and they have been shown to reduce variability in clinical practice. Other advantages include decision-making support for the clinician and optimization of resources, generating savings in time and costs.
ConclusionsA huge amount of scientific and medical information is now available. However, there is an inherent problem in such a volume of data: selective recovery and interpretation are practically impossible for a professional using classical methods. In this setting, the use of bioinformatics tools such as text or data mining acquires fundamental relevance. These tools, which already play a significant role in other areas of biomedical knowledge, have recently begun to be used in respiratory medicine. The most obvious areas for the medical application of text mining (both in research and in the clinic) are the integration and transfer of advances made in the most basic sciences, and a better understanding of the diagnostic processes, severity classifications and determination of disease prognosis. Text mining may also be useful for generating predictive outcomes models, creating intelligent alert systems and supporting the clinician in the decision-making process.
Conflict of InterestThe authors declare no conflict of interest.
The authors thank Dr Ferran Sanz and Dr Laura Furlong of the Research in Informatics Program (GRIB, IMIM – UPF), for their guidance and advice in the writing of this manuscript.
Please cite this article as: Piedra D, Ferrer A, Gea J. Minería de textos y medicina: utilidad en las enfermedades respiratorias. Arch Bronconeumol. 2014;50:113–119.