High-altitude pulmonary edema (HAPE) is a kind of non-cardiogenic edema with high incidence and life-threatening. This study was designed to explore the association of LINC-PINT and LINC00599 polymorphisms with HAPE susceptibility.
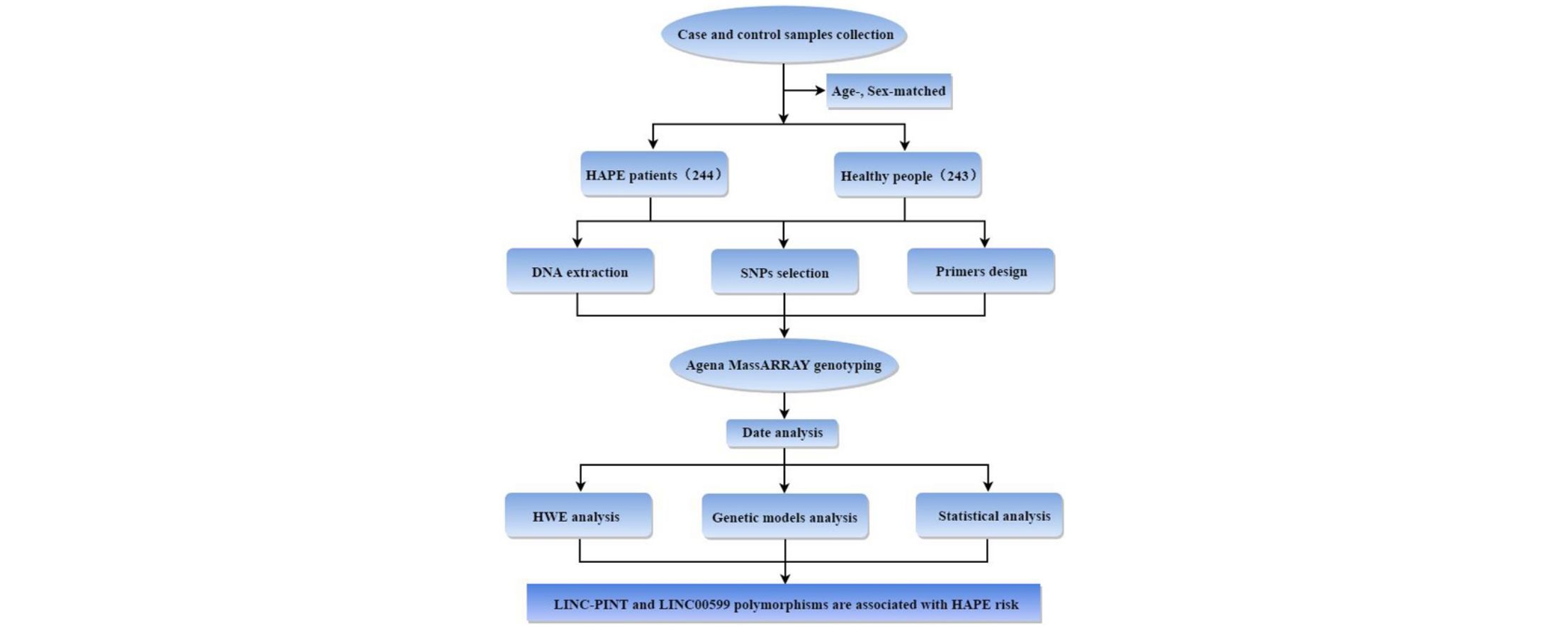
MethodsThis study included 244 HAPE patients and 243 age-, sex-matched healthy controls from the Chinese population. The genotypes of polymorphisms were detected using the Agena MassARRAY. The relationship between polymorphisms and HAPE risk was evaluated using a χ2 test with an odds ratio (OR) and 95% confidence intervals (CIs) in multiple genetic models.
ResultsWe observe a significant association between the rs157928 and decreased HAPE risk in genotype model (OR=0.65, 95% CI=0.43–0.98, p=0.038). The subgroup analysis results indicated that rs2272026 was associated with a decreased risk of HAPE in younger patients with age ≤32 (codominant model: p=0.006; recessive model: p=0.005 additive model: p=0.018; and allele model: p=0.012; rs72625676, codominant model: p=0.038; recessive model: p=0.037). Among patients older than 32 years, there was a significantly increased risk of HAPE associated with the rs2272026 and rs1962430 (rs2272026: genotype model: p=0.049; recessive model: p=0.029; rs1962430: genotype model: p=0.024; recessive model: p=0.020). Nevertheless, rs157928 had relationship with significantly reducing the risk of HAPE in the genotype model (p=0.018).
ConclusionOur study suggests that LINC-PINT and LINC00599 polymorphisms are associated with HAPE susceptibility in Chinese population.
El edema pulmonar de altitud (EPA) es un tipo de edema no cardiogénico con una incidencia alta y es potencialmente mortal. Este estudio se diseñó para explorar la asociación entre los polimorfismos de LINC-PINT y LINC00599 y la susceptibilidad al EPA.
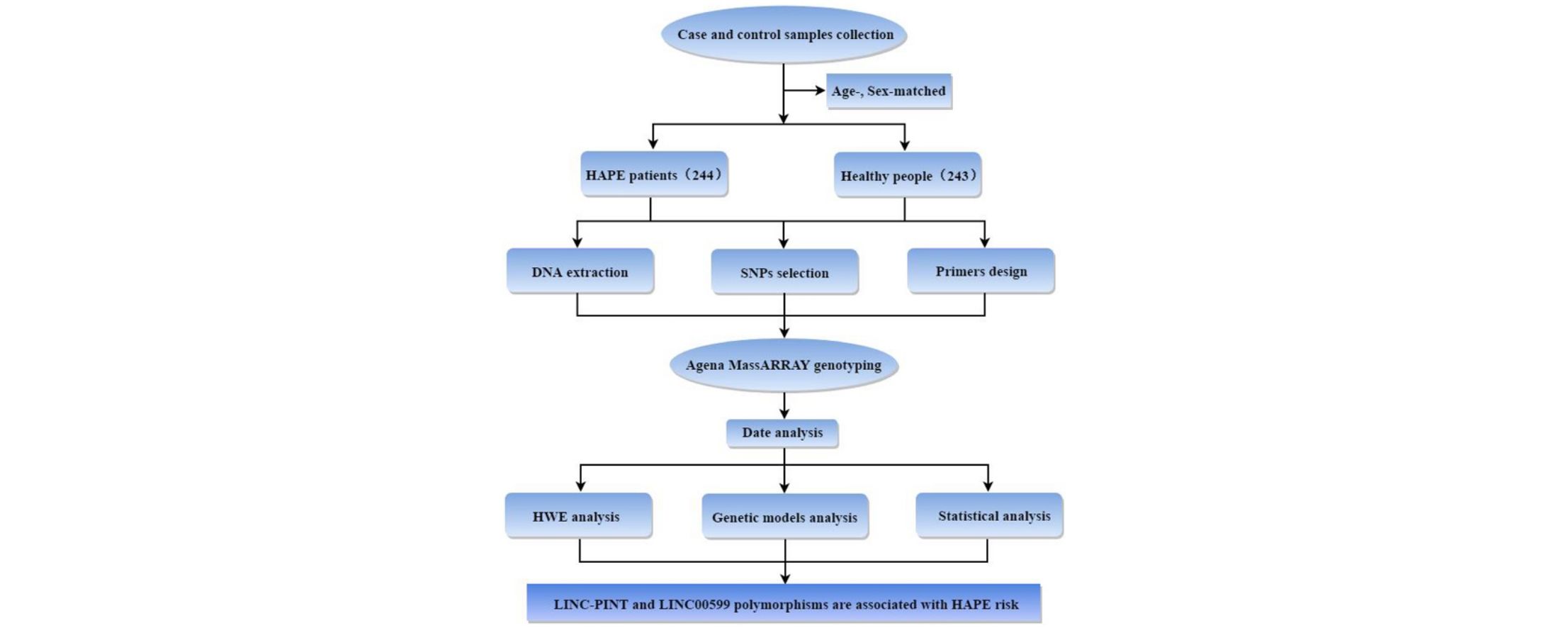
MétodosEste estudio incluyó a 244 pacientes con EPA y 243 controles sanos de la misma edad y sexo, todos de origen chino. Los genotipos de los polimorfismos se detectaron utilizando el MassARRAY de Agena™. La relación entre los polimorfismos y el riesgo de EPA se evaluó utilizando el test de la χ2 con el odds ratio (OR) e intervalos de confianza (IC) del 95% en múltiples modelos genéticos.
ResultadosObservamos una asociación significativa entre el rs157928 y una disminución del riesgo de EPA en el modelo genotípico (OR=0,65, IC del 95%=0,43-0,98, p=0,038). Los resultados de los análisis por subgrupos indicaron que el rs2272026 se asociaba a una disminución del riesgo de EPA en pacientes jóvenes de ≤ 32 años (modelo codominante: p=0,006; modelo recesivo: p=0,005; modelo aditivo: p=0,018; y modelo por alelos: p=0,012 para rs72625676, modelo concomitante: p=0,038; modelo recesivo: p=0,037). Entre los pacientes mayores de 32 años, se encontró un riesgo aumentado de EPA de manera significativa asociado a los rs2272026 y rs1962430 (rs2272026: modelo genotípico: p=0,049; modelo recesivo: p=0,029; rs1962430: modelo genotípico: p=0,024; modelo recesivo: p=0,020). Sin embargo, el rs157928 tenía relación con una reducción significativa del riesgo de EPA en el modelo genotípico (p=0,018).
ConclusiónNuestro estudio sugiere que los polimorfismos de LINC-PINT y LINC00599 están asociados a la susceptibilidad de EPA en la población china.
High altitude pulmonary edema (HAPE) is a lethal plateau acute idiopathic disease. It occurs often in two forms. The first form usually occurs in un-acclimatized healthy lowlanders who first arrive at the plateau. The second form, also called re-entry HAPE, occurs among high-altitude residents who return after a low-altitude sojourn.1 HAPE is one of the leading causes of death in altitude sickness, which seriously endangers people's health and quality of life.2,3 At present, the pathogenesis is thought to be related to pulmonary hypertension, increased vascular permeability, stress response, and inflammatory response triggered by hypoxia in the high altitude area. However, under the same hypoxic conditions, some people are more prone to HAPE, so the genetic factor is an un-negligible factor.4 And more and more studies have shown the importance of genetic factors in HAPE.5–7 To date, polymorphisms of NOS3,8AGTR1,9ADRB2,10HLA11 and other genes has been proved to be positively correlated with the risk of HAPE. Previously, Japanese scholar found that the expression level of HLA genes was significantly increased in HAPE patients, indicating that extensive studies on genetic susceptibility have laid an important foundation for revealing the pathogenesis of HAPE and pointed out a new direction. However, the genetic study of HAPE is relatively lacking, and more susceptibility genes and their functional mechanisms need to be further explored.12
Long non-coding RNAs (lncRNAs) are non-protein coding RNA segments with a length of greater than 200 nucleotides. LncRNA has many biological functions, such as cell overgrowth, cell differentiation and apoptosis, etc. Recent studies have found that some lncRNAs can affect the proliferation of pulmonary artery smooth muscle cells, especially in patients with hypoxia-induced pulmonary hypertension,13–15 and we know the pulmonary hypertension can easily develop into HAPE. By reading the relevant literature, we found two HAPE-related lncRNAs as research objects. The lncRNA LINC-PINT differentially expressed in the lungs and inhibited progression of tumor cells.16 We suspect that it may be involved in the development and progression of lung-related diseases. And another lncRNA LINC00599 was identified as a promising target for treating cardiovascular disorders.17 Therefore, we suspect that the LINC00599 may also be involved in pulmonary arterial vascular diseases, including HAPE. Combined with the above, we hypothesized that the polymorphism of these two lncRNAs play important role in HAPE.
Therefore, in this study, we will conduct a case–control study and identify the associations between HAPE and nine susceptible SNPs in the LINC-PINT and LINC00599 to further clarify their potential roles in HAPE risk in the Chinese population. The study aims to identify a positive finding for the early prevention of HAPE.
Materials and methodsStudy subjectsA case–control study was performed with 244 patients with HAPE and 243 healthy controls randomly recruited from 2017 to 2018 in the Affiliated Hospital of Xizang Minzu University. All of the subjects were of genetically unrelated Chinese ancestry. All patients and healthy people were previously resident in low-altitude areas below 2500m, and moved to high altitude areas above 3000m due to work or travel. The patients were diagnosed with HAPE based on the diagnostic criteria (GBZ 92–2008). Patients with a prior cardiopulmonary history were excluded by standardized questionnaires and clinical examination. The healthy controls and patients were from the same area, matched in age, sex and living environment. The controls had lived at high altitudes for at least three months, and had not suffered from HAPE or any other mountain sickness. The controls with other serious cardiopulmonary diseases or cancers were excluded. This study was approved by the Ethics Committee of the Xizang Minzu University (Number: 201702), and every subject signed written consent.
Genotyping assayFasting peripheral blood of all participants were collected in anticoagulant tubes and stored at −80°C. The whole blood genomic DNA extraction kit (GoldMag Co. Ltd., Xi’an, China) was used to extract DNA in accordance with the manufacturer's protocol, and the DNA content was measured by spectrometry (NanoDrop 2000 spectrophotometer, Thermo Scientific, Waltham, Massachusetts, USA). Nine single nucleotide polymorphisms (SNPs) in LINC-PINT and LINC00599 with a minor allele frequency (MAF) >5% of the 1000 Genomes Project data (http://www.internationalgenome.org/) were selected in the present study. Agena MassARRAY Assay Design Software (version 3.0, Agena Bioscience, San Diego, USA) was used to design multiplexed SNP MassEXTEND assay. And Agena MassARRAY RS1000 was used to detect SNP genotyping.18,19 Data were analyzed with Agena Typer Software (version 4.0, Agena Bioscience, San Diego, CA, USA).
Statistical analysisSPSS software (version 22.0, Chicago, IL, USA) was used for statistical analyses of data. The independent sample t-test or χ2 test was used to examine the differences of basic parameters between the two groups. Hardy–Weinberg equilibrium as well as the differences in allele frequencies between cases and controls was examined by χ2 test. The HAPE risk associated with genotypes was estimated by odds ratios (ORs) with 95% confidence intervals (CIs) for five different genetic models. For all test, a two-tailed p-value <0.05 was considered statistically significant. HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) was conducted to predict the potential functions of the SNPs.
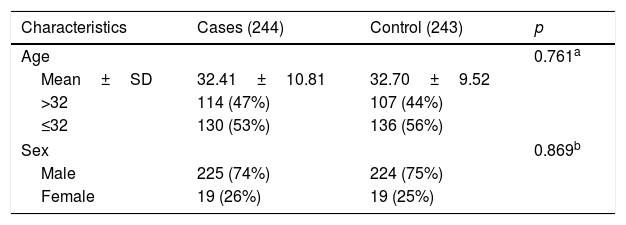
ResultsThis study included 244 patients with HAPE and 243 healthy controls. The clinical and demographic characteristic of patients and controls are shown in Table 1. The cases and controls were matched by age and sex (p>0.05). The mean age of cases and controls are 32.41±10.81 and 32.70±9.52 respectively. The detailed information of nine SNPs of LINC-PINT and LINC00599 was listed in Table 2. For all SNPs, the MAFs were greater than 5% and the observed genotype frequencies complied with HWE in the control group. In addition, we annotate the functional elements of these selected SNPs using HaploRegv4.1. Through the prediction of this online software, we found that candidate SNPs may have the following functions: 1. These SNPs may promote histone marked. 2. These SNPs may enhance histone marked. 3. They may be involved in DNA enzyme synthesis. 4. They may work by binding directly to proteins. 5. These SNPs may alter the conformation of host genes-encoding proteins. 6. Finally, they may affect the level of genes expression. However, the exact function needs further experimental verification.
Basic information of candidate single nucleotide polymorphisms (SNPs) in the study.
| Gene | SNP_ID | Role | Position | Case (244) | Control (243) | p-HWE | HaploReg | ||
|---|---|---|---|---|---|---|---|---|---|
| MA | MAF | MA | MAF | ||||||
| LINC-PINT | rs157916 | Intronic | 130884634 | G(206) | 0.422 | G(189) | 0.390 | 0.686 | Promoter histone marks, Enhancer histone marks, DNAse, Proteins bound, Motifs changed, Selected eQTL hits |
| rs16873842 | Intronic | 130885731 | A(75) | 0.154 | A(78) | 0.160 | 0.349 | Enhancer histone marks, DNAse, Motifs changed, Selected eQTL hits | |
| rs7801029 | Intronic | 130890856 | G(109) | 0.223 | G(102) | 0.210 | 0.437 | Promoter histone marks, Enhancer histone marks, DNAse, Proteins bound, Motifs changed | |
| rs7781295 | Intronic | 130894403 | A(90) | 0.185 | A(85) | 0.176 | 0.375 | Promoter histone marks, Enhancer histone marks, DNAse, Proteins bound, Motifs changed, Selected eQTL hits | |
| rs28662387 | Intronic | 130895393 | A(26) | 0.053 | A(27) | 0.056 | 0.534 | Enhancer histone marks, DNAse, Proteins bound, Motifs changed | |
| rs157928 | Intronic | 130896599 | C(210) | 0.430 | C(210) | 0.432 | 0.192 | Enhancer histone marks, DNAse, Proteins bound, Motifs changed | |
| LINC00599 | rs2272026 | Exonic | 9900090 | C(186) | 0.381 | C(196) | 0.410 | 0.595 | Promoter histone marks, Enhancer histone marks, DNAse, Motifs changed, Selected eQTL hits |
| rs1962430 | Exonic | 9902110 | T(161) | 0.330 | T(156) | 0.321 | 0.302 | Promoter histone marks, Enhancer histone marks, DNAse, Selected eQTL hits | |
| rs531564 | Exonic | 9903189 | C(62) | 0.127 | C(69) | 0.142 | 0.796 | Promoter histone marks, Enhancer histone marks, DNAse, Motifs changed, GRASP QTL hits | |
SNPs, single nucleotide polymorphism; MA, minor allele; MAF, minor allele frequency; HWE, Hardy–Weinberg equilibrium.
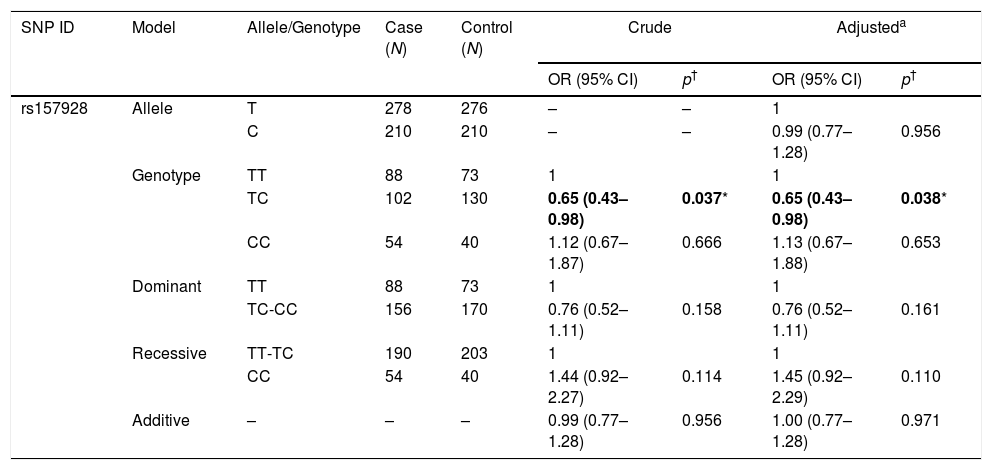
The association between rs157928 of LINC-PINT and risk of HAPE is shown in Table 3. We evaluated this association in five genetic models used χ2 test and odds ratios (ORs) with logistic regression adjusted for age and sex. We observe a significant association between the rs157928 and decreased HAPE risk in genotype model (TC: OR=0.65, 95% CI=0.43–0.98, p=0.038).
Analysis of association between rs157928 of LINC-PINT and risk of HAPE.
| SNP ID | Model | Allele/Genotype | Case (N) | Control (N) | Crude | Adjusteda | ||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p† | OR (95% CI) | p† | |||||
| rs157928 | Allele | T | 278 | 276 | – | – | 1 | |
| C | 210 | 210 | – | – | 0.99 (0.77–1.28) | 0.956 | ||
| Genotype | TT | 88 | 73 | 1 | 1 | |||
| TC | 102 | 130 | 0.65 (0.43–0.98) | 0.037* | 0.65 (0.43–0.98) | 0.038* | ||
| CC | 54 | 40 | 1.12 (0.67–1.87) | 0.666 | 1.13 (0.67–1.88) | 0.653 | ||
| Dominant | TT | 88 | 73 | 1 | 1 | |||
| TC-CC | 156 | 170 | 0.76 (0.52–1.11) | 0.158 | 0.76 (0.52–1.11) | 0.161 | ||
| Recessive | TT-TC | 190 | 203 | 1 | 1 | |||
| CC | 54 | 40 | 1.44 (0.92–2.27) | 0.114 | 1.45 (0.92–2.29) | 0.110 | ||
| Additive | – | – | – | 0.99 (0.77–1.28) | 0.956 | 1.00 (0.77–1.28) | 0.971 | |
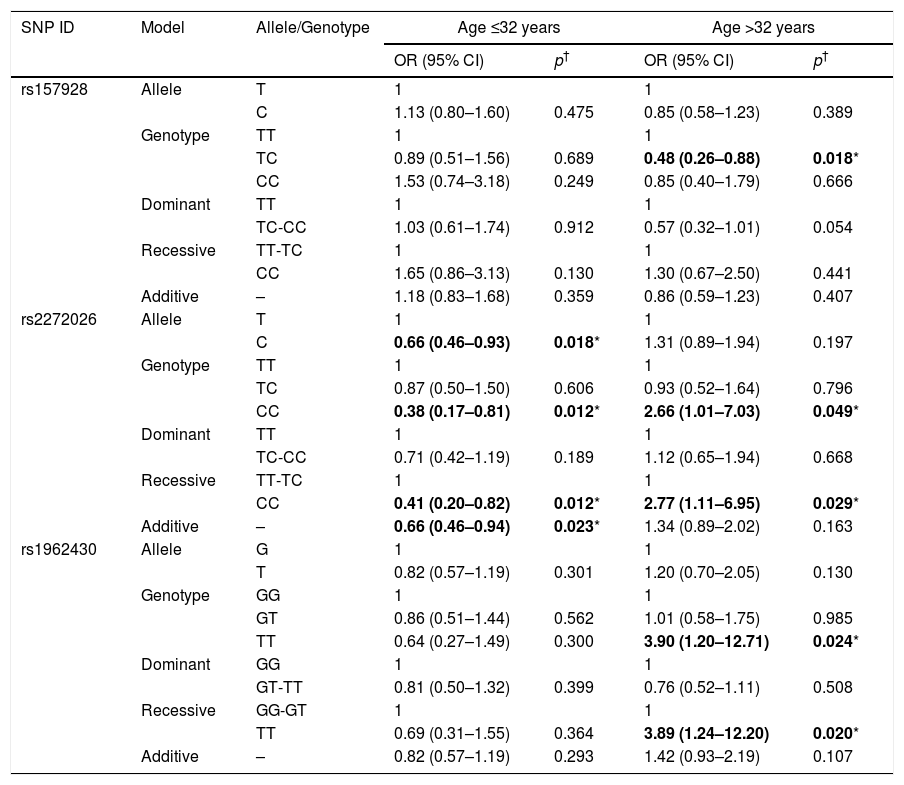
Then we performed a subgroup analysis regarding the effect of rs157928, rs2272026 and rs1962430 polymorphisms on HAPE according to age is displayed in Table 4. The results indicated that rs2272026 was associated with a decreased risk of HAPE in the people younger than 32 years old (allele model: C: OR=0.66, 95% CI=0.46–0.93, p=0.018; genotype model: CC; OR=0.38, 95% CI=0.17–0.81, p=0.012; recessive model: CC: OR=0.41, 95% CI=0.20–0.82, p=0.012; additive model: OR=0.66, 95% CI=0.46–0.94, p=0.023). In the subgroup older than 32 years, we found a significantly increased risk of HAPE associated with the variant genotypes rs2272026-CC and rs1962430-TT in two models (rs2272026: genotype model: OR=2.66, p=0.049; recessive model: OR=2.77, p=0.029; rs1962430: genotype model: OR=3.90, p=0.024; recessive model: OR=3.89, p=0.020). Nevertheless, rs157928-TC had a relationship with significantly decreasing the risk of HAPE in the genotype model (OR=0.48, 95% CI=0.26–0.88, p=0.018).
Stratified analysis of polymorphisms in LINC-PINT and LINC00599 on HAPE risk by age.
| SNP ID | Model | Allele/Genotype | Age ≤32 years | Age >32 years | ||
|---|---|---|---|---|---|---|
| OR (95% CI) | p† | OR (95% CI) | p† | |||
| rs157928 | Allele | T | 1 | 1 | ||
| C | 1.13 (0.80–1.60) | 0.475 | 0.85 (0.58–1.23) | 0.389 | ||
| Genotype | TT | 1 | 1 | |||
| TC | 0.89 (0.51–1.56) | 0.689 | 0.48 (0.26–0.88) | 0.018* | ||
| CC | 1.53 (0.74–3.18) | 0.249 | 0.85 (0.40–1.79) | 0.666 | ||
| Dominant | TT | 1 | 1 | |||
| TC-CC | 1.03 (0.61–1.74) | 0.912 | 0.57 (0.32–1.01) | 0.054 | ||
| Recessive | TT-TC | 1 | 1 | |||
| CC | 1.65 (0.86–3.13) | 0.130 | 1.30 (0.67–2.50) | 0.441 | ||
| Additive | – | 1.18 (0.83–1.68) | 0.359 | 0.86 (0.59–1.23) | 0.407 | |
| rs2272026 | Allele | T | 1 | 1 | ||
| C | 0.66 (0.46–0.93) | 0.018* | 1.31 (0.89–1.94) | 0.197 | ||
| Genotype | TT | 1 | 1 | |||
| TC | 0.87 (0.50–1.50) | 0.606 | 0.93 (0.52–1.64) | 0.796 | ||
| CC | 0.38 (0.17–0.81) | 0.012* | 2.66 (1.01–7.03) | 0.049* | ||
| Dominant | TT | 1 | 1 | |||
| TC-CC | 0.71 (0.42–1.19) | 0.189 | 1.12 (0.65–1.94) | 0.668 | ||
| Recessive | TT-TC | 1 | 1 | |||
| CC | 0.41 (0.20–0.82) | 0.012* | 2.77 (1.11–6.95) | 0.029* | ||
| Additive | – | 0.66 (0.46–0.94) | 0.023* | 1.34 (0.89–2.02) | 0.163 | |
| rs1962430 | Allele | G | 1 | 1 | ||
| T | 0.82 (0.57–1.19) | 0.301 | 1.20 (0.70–2.05) | 0.130 | ||
| Genotype | GG | 1 | 1 | |||
| GT | 0.86 (0.51–1.44) | 0.562 | 1.01 (0.58–1.75) | 0.985 | ||
| TT | 0.64 (0.27–1.49) | 0.300 | 3.90 (1.20–12.71) | 0.024* | ||
| Dominant | GG | 1 | 1 | |||
| GT-TT | 0.81 (0.50–1.32) | 0.399 | 0.76 (0.52–1.11) | 0.508 | ||
| Recessive | GG-GT | 1 | 1 | |||
| TT | 0.69 (0.31–1.55) | 0.364 | 3.89 (1.24–12.20) | 0.020* | ||
| Additive | – | 0.82 (0.57–1.19) | 0.293 | 1.42 (0.93–2.19) | 0.107 | |
In this case–control study, we investigated the association between polymorphisms of LINC-PINT, LINC00599 and risk of HAPE. Rs157928 was associated with decreased risk of HAPE, whereas rs1962430 was associated with significantly increased risk. Additionally, the effect of rs2272026 is reversed in different age groups. To our knowledge, this is the first study to report the effect of these SNPs in HAPE individuals.
Long non-coding RNAs are a vital class of RNAs, which function in regulating gene expression, and are implicated in multiple cancers and cardiovascular diseases. In recent years, it has been found that some LncRNAs are involved in the pathogenesis of atherosclerosis, pulmonary arterial hypertension, especially hypoxic pulmonary hypertension. LncRNAs are abundant in vascular cells and plays an important role in the contractility of smooth muscle cells. In addition, the proliferation and apoptosis of vascular muscle cells are regulated by lncRNAs.15,20,21
In a previous study, the authors found that genetic variations in lncRNAs can influence their expression or functions and were associated with diseases initiation and progression. Rs619586 in the lncRNA MALAT1 was associated with a reduced risk of pulmonary arterial hypertension. Furthermore, genetic variations may affect MALAT1's function in promoting cell proliferation and migration.21 In our study, two new lncRNAs (LINC-PINT and LINC00599) were found to affect the risk of developing HAPE. LINC-PINT-rs157928 was identified as a protective factor in HAPE. In non-small cell lung cancer, over-expression of LINC-PINT has been shown to inhibit the proliferation, cycle progression, migration and invasion of cancer cells, thereby inhibiting the occurrence of non-small cell lung tumors.16 From the above, we hypothesized that rs157928 was a meaningful site, which might affect the function of LINC-PINT, and thus the susceptibility to pulmonary edema at altitude.
The expression of LINC00599 was first detected during retinal development in mouse.22 Because the vascular systems of the eyes and heart share many features, LINC00599 was soon found to be involved in the development of cardiovascular diseases, such as atherosclerosis.20,23 In addition, the function of LINC00599 in human cancer is gradually attracting the attention of scientists. In prostate cancer, LINC00599 regulates the proliferation and invasion of prostate cancer cells and predicts tumor progression and poor survival in patients. In glioblastoma, LINC00599 was down-regulated in tumor cell lines, played an anti-cancer role in regulating the proliferation and apoptosis of glioma cells, and was related to the prognosis of glioma.24,25 However, the function of LINC00599 in lung-related diseases is unclear. In this study, we found that two SNPs (rs2272026 and rs1962430) of LINC00599 affected the risk of HAPE. We hope LncRNA is an ideal target for the treatment of HAPE and related cardiopulmonary and vascular diseases.
Several limitations of this study should be pointed out. First, we only genotyped nine SNPs in the LINC-PINT and LINC00599 in this study. More other SNPs should be examined in future studies. Second, our study enrolled only a moderate number of subjects, and we needed to conduct a larger survey to confirm our findings. In particular, the results of stratified analysis need to be verified in experiments with larger samples.
ConclusionIn summary, this case–control study indicates that the LINC-PINT and LINC00599 polymorphisms are associated with HAPE susceptibility in the Chinese population. Further functional studies and large population-based prospective studies are being planned to provide accurate evidence about the influence of genetic variants on HAPE.
AuthorshipConceptualization, Dongya Yuan, Xue He and Tianbo Jin; methodology, Xue He; software, Jianwen Zheng; validation, Yongjun He, Yuhe Wang and Li Wang; data curation, Xue He; writing original draft preparation, Mei Bai; writing, review and editing, Xue He; visualization, Xue He and Jianwen Zheng.
Conflict of interestAll authors declare that they have no conflict of interests.
We thank all participants for taking part in this study.
This work was supported by the National Natural Science Foundation of China [81860330] and Major Training Program of Xizang Minzu Univerisity [18MDZ04].