Lung cancer is a major public health problem, as the second causes of cancer related death worldwide, with relatively low survival rates, and accessible drug resistance. Long non-coding RNAs (LncRNAs) have been identified as activator in lung cancer with elusive mechanisms. We aimed to detect the regulation of LncRNA MALAT1 in the proliferation and gefitinib resistance in lung cancer cells.
MethodsMALAT1 in A549 and HCC 1299 human lung adenocarcinoma cell lines was silenced by shRNA or overexpressed using plasmid, and the cell viability and cell proliferation were evaluated by MTT assay and soft agar colony formation assay. RNA levels were detected by RT-PCR, and the protein expression was measured by western blot. The binding between MALAT1 and miR-200a was validated by luciferase reporter assays using pSi-Chech 2 vectors.
ResultsThe cell viability and proliferation of A549 cells transfected with MALAT1 shRNA were significantly lower than the control. The MALAT1 expression in gefitinib resistant A549 cells was upregulated. miR-200a significantly inhibited the fluorescence of pSi-Check 2 vector with MALAT1 gene, suggesting the direct binding between MALAT1 and miR-200a. In addition, LncRNA MALAT1 promotes ZEB1 expression in A549 cells.
ConclusionOur study showed that MALAT1 promoted the proliferation and gefitinib resistance of lung cancer cells by sponging miR-200a, which regulates expression of ZEB1 in the A549 cells. This MALAT1/miR-200a axis could serve as new therapeutic target for lung cancer treatment.
El cáncer de pulmón es un importante problema de salud pública. Constituye la segunda causa de muerte por cáncer en el mundo y se asocia a tasas de supervivencia relativamente bajas y a resistencia a fármacos accesibles. Los RNA largos no-codificantes (lncRNA) se han identificado como activadores en el cáncer de pulmón con mecanismos aún no esclarecidos. El objetivo de este estudio fue detectar la regulación del LncRNA MALAT1 en la proliferación y la resistencia de las células tumorales de pulmón.
MétodosLa expresión de MALAT1 se silenció con un shRNA o se sobreexpresó mediante el uso de un plásmido en las líneas celulares de adenocarcinoma pulmonar humano A459 y HCC. La viabilidad celular y la proliferación se evaluaron mediante un ensayo MTT y el ensayo de formación de colonias en agar blando. Los niveles de RNA se detectaron con RT-PCR y los niveles de proteína se midieron con Western Blot. La unión entre MALAT1 y miR-200a se validó gracias a un ensayo de luciferasa utilizando vectores pSi-Chech 2.
ResultadosLa viabilidad y la proliferación de las células A549 transfectadas con el shRNA contra MALAT1 resultaron significativamente más bajas que en el control. Se produjo un incremento en la expresión de MALAT1 en las células A549 resistentes a gefitinib. MiR-200a inhibió significativamente la fluorescencia del vector pSi-Check 2 que contenía el gen MALAT1, sugiriendo la unión directa entre MALAT1 y miR-200a. Además, el LncRNA de MALAT1 promovió la expresión de ZEB1 en las células A549.
ConclusiónNuestro estudio demuestra que MALAT1 promovió la proliferación y resistencia a gefitinib en células tumorales de pulmón actuando como “esponja” de miR-200a, que regula la expresión de ZEB1 en células A549.
Lung cancer is a major public health problem, being considered as the second causes of cancer related death worldwide.1 Lung cancer can be categorized into two broad types including primary and secondary cancers. The former is characterized by the lung tissues originated carcinoma cells, while the latter is induced by the carcinoma cells metastasized to the lung tissues.2 Owing to the improvements in the chemotherapy and surgical techniques, the survival rate of lung cancer has largely increased compared to several decades ago, while the relative 5-year survival rate is 55% for localized cases. However, the survival rates for regional disease and distant stage disease are only approximately 27% and 4%, respectively.3 To overcome this, researchers have been working hard to develop new therapeutic strategies for lung cancer treatment.
It has been recently demonstrated that the epidermal growth factor receptor (EGFR) signaling is an attractive target for cancer therapy, due to its frequent overexpression or aberrantly activation in non-small cell lung cancer (NSCLC), which accounts for approximately 80% of all lung cancer cases.4 Gefitinib is a selective inhibitor of epidermal growth factor, which plays a critical role in the cell proliferation, apoptosis, and angiogenesis.5 Since developed, gefitinib has been widely used as first-line treatment of advanced NSCLC, in chemotherapy-naive patients, or as later-line treatment after chemotherapy failure.6 Unfortunately, in spite of the brilliant initial responses to EGFR-tyrosine kinase inhibitors, most patients with lung adenocarcinoma eventually develop resistance to anti-EGFR agents within 12 months.7 Currently, accumulating studies have revealed different mechanisms such as HER3-related activation of PI3K/AKT pathways,8 abnormal activation of c-Met,9 and increased expression of DDX1710 in NSCLC cells have been reported to be responsible for the development of gefitinib resistance, although the large part of the mechanisms underlying the gefitinib resistance to EGFR-TKI are still not completely understood.
Long non-coding RNAs (LncRNAs), consist of >200 nucleotides, are a diverse class of RNAs that engage in various biological processes, such as cell proliferation, survival, apoptosis, energy metabolism, and tumourigenesis.11,12 The downstream effectors of LncRNAs include microRNAs (miRs) and proteins. In the recent years, a great number of LncRNAs have been identified as activator or suppressor in various human cancer types, including lung.13 The LncRNA metastasis associated lung adenocarcinoma transcript 1 (MALAT1), which is a highly conserved nuclear LncRNA has been reported as a predictive marker for metastasis development in lung cancer.14 However, it remains unclear how MALAT1 regulate the tumor growth.
In the present study, we aimed to detect the regulation of LncRNA MALAT1 in the proliferation of lung cancer, especially the role of MALAT1 on the gefitinib resistance in lung cancer cells. The downstream microRNA and protein targets of LncRNA MALAT1 were also investigated.
Materials and MethodsA549 and HCC 1299 Lung Cancer Cell Line CultureThe A549 and HCC 1299 human lung adenocarcinoma cell lines were obtained from Shanghai Liangtai Biotechnology (Shanghai, China). Cells were cultivated in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS), 0.2mM glutamine, 100U/mL penicillin, and 100μg/mL streptomycin (Gibco, Grand Island, NY, USA). All of the materials were obtained upon the approval of Material Transfer Agreement (MTA) between institutional ethical committees.
RNA Extraction and Quantitative Real Time PCRRNA was extracted from the cultured cells using Trizol reagent (Thermo Fisher Scientific, Gaithersburg, MD, USA). The isolated total RNA was reverse transcribed into cDNA using the PrimeScript RT Reagent Kit (Takara Bio., Dalian, China) according to the manufacturer's instruction. To detect lncRNA MALAT1 expression, RT-PCR was performed using the SYBR-Green qPCR mix (Bio-rad, Hercules, CA, USA) according to the manufacturer's protocol.
The primer sequences used were as follows: MALAT1 forward, 5′-GGTAACGATGGTGTCGAGGTC-3′ and reverse, 5′-CCAGCATTACAGTTCTTGAACATG-3′. β-Actin was used as internal control.
Cell Viability Detection by MTT AssayTo determine the viability of tumor cells, cells were seeded into 96-well plate at 500cells/well and cultured in DMEM supplemented with 10% FBS. Cells viability was measured using MTT reagent (Sigma-Aldrich, St. Louis, MO) at 5mg/ml in PBS. Briefly, cells were incubated with fresh medium and diluted MTT (1:10, 10% MTT), for 4h at 37°C. Further, the formazan crystals were dissolved in 200μl of dimethyl sulfoxide solution, and the absorbance was quantified at 570nm using microplate reader (BioTek Instruments, VT, USA).
The Soft Agar Colony Formation AssayFollowing the plating of bottom layer of agar, the A549 cells were harvested and diluted in culture medium, then mixed 0.6% agar and cell suspension in a 1:1 ratio, and plated the upper layer of agar containing cells at 5000cells/well. The agar plates were cultures at 37°C for 20 days, and the colonies obtained were stained by 200μl of nitroblue tetrazolium chloride solution per well and incubated overnight at 37°C. The colonies were counted and compared among groups.
The Induction of Gefitinib Resistant A549 CellsThe A549 cells were incubated with gefitinib at a starting concentration of 0.05mM, which was gradually increased to 1mM, for 6 continuous months. The cells were incubated with gefitinib at 0, 0.005, 0.01, 0.05, 0.1, 0.5 and 1mM, and the ones tolerate at 1mM of gefitinib were identified as gefitinib resistant A549 cells (A549 GR). The doses were chosen based on the published paper.15
Luciferase Reporter AssaysThe binding domain of microRNA on MALAT1 was predicted using bioinformatics tool starBase v2.0 (http://starbase.sysu.edu.cn/panCancer.php). The reporter vector pSi-Chech 2-MALAT1 was constructed using pSi-Chech 2 vector bone. A549 cells were co-transfected with 0.5μg of the reporter vector. In addition, 1μg of MALAT1 expression plasmid or the control vector (Shanghai Genepharma Inc., China) were transfected into A549 cells using Lipofectamine 2000 (ThermoFisher Scientific). Forty-eight hours post transfection, A549 cells were harvested and luciferase activity was measured using dual-luciferase reporter assay (Promega, Madison, WI, USA).
Protein Expression Assessment by Western BlotTotal 20μg of protein from A549 cell lysates was separated by 12% SDS-PAGE and transferred to PVDF membrane (Bio-rad, Hercules, CA, USA). The membranes were blocked for 2h at room temperature with 5% non-fat milk in TBST (TBS, 0.1% Tween 20). The membrane was further incubated overnight at 4°C with primary antibody against ZEB 1 and GADPH (Cell Signaling, MA, USA). Membranes were washed five times and incubated with HRP conjugated secondary antibodies for 1h. All membranes were detected using enhanced chemiluminescence substrate (Thermo Fisher Scientific).
Statistical AnalysisAll statistical analyses were performed using the SPSS 11.2 software. All experimental data are reported as the mean±standard deviation (SD). The Student's t-test or ANOVA analysis was used in the analysis. Comparison results with P value as *P<.05 were considered statistically significant.
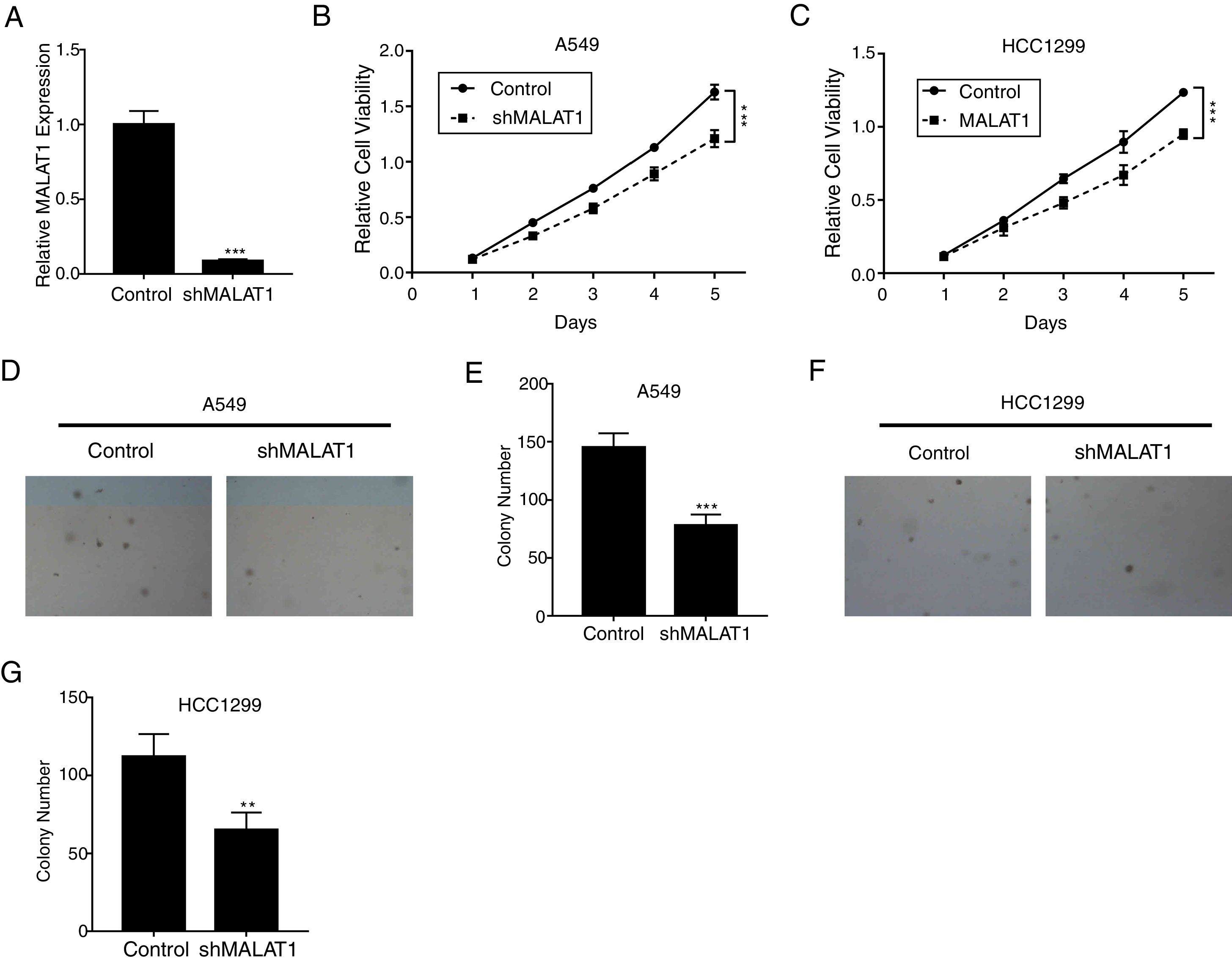
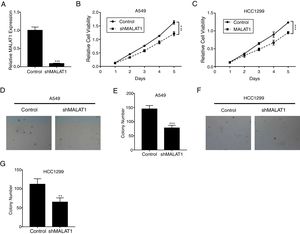
ResultsLncRNA MALAT1 Promotes Lung Cancer Cells ProliferationTo determine the effects of LncRNA MALAT1 on the cell proliferation of cancer cells, MALAT1and control shRNA were used to transfect A549 and HCC1299 cells. As shown in Fig. 1A, the relative MALAT1 expression in shMALAT1 transfected cells were significantly lower than the expression in the control cells (P<.001), indicating the effective knock-down of MALAT1 gene. From Fig. 1B and C, we observed that the relative cell viabilities of shMALAT1 transfected A549 and HCC1299 cells were significantly lower than the control cells in a time-dependent manner (P<.001). Further, we used soft agar colony formation assay to detect the tumor cell proliferation. As demonstrated in Fig. 1D and E, the A549 cells transfected with shMALAT1 showed significantly lower quantity of cells on the layer of agar plate (P<.001). Similar results were observed for the colony numbers of HCC1299 cells on agar plate (Fig. 1F and G, P<.01).
LncRNA MALAT1 promotes lung cancer cells proliferation. (A) The mRNA levels of LncRNA MALAT1 in A549 cells transfected with MALAT1 shRNA or control vector were determined by qPCR. (B) The proliferation of A549 cells transfected with MALAT1 shRNA was determined by MTT assay. (C) The proliferation of HCC1299 cells transfected with MALAT1 shRNA was determined by MTT assay. (D) The proliferation of A549 cells transfected with MALAT1 shRNA was determined by soft agar colony formation assay. (E) The statistical results of soft agar colony formation assay. (F) The proliferation of HCC1299 cells transfected with MALAT1 shRNA was determined by soft agar colony formation assay. (G) The statistical results of soft agar colony formation assay. *** P < .001.
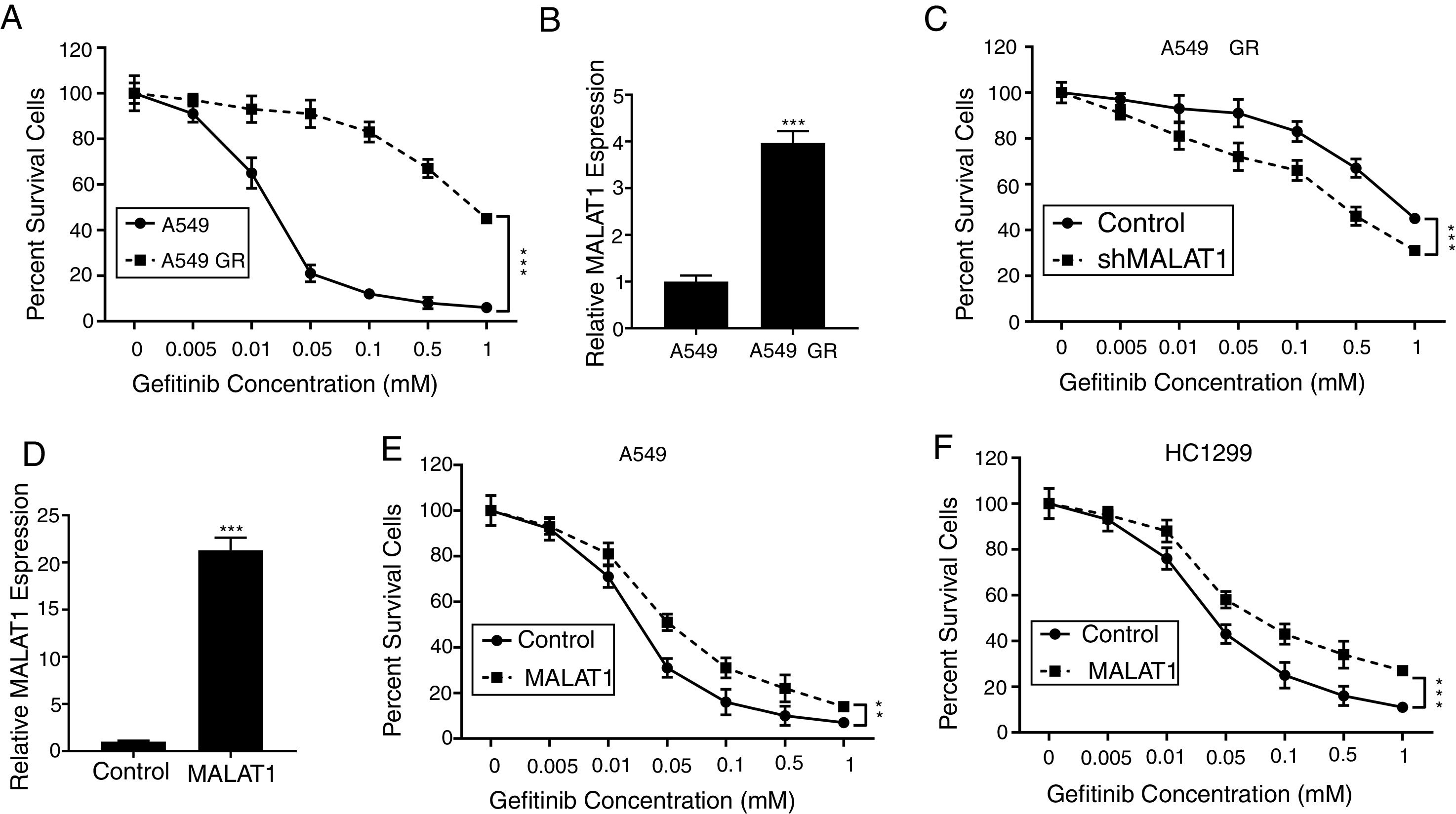
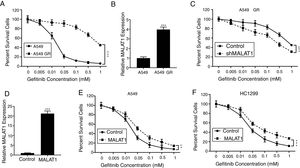
To test the effects of LncRNA MALAT1 on the gefitinib resistance in lung cancer cells, we firstly established the gefitinib resistant A549 cells (A549 GR) by culturing the cells with gefitinib at increasing concentrations. As seen in Fig. 2A, we saw percentage of survival A549 GR cells retained 40%, whereas the percentage of survival in normal A549 cells dropped to less than 5% (P<.001). Strikingly, we found the relative expression of MALAT1in A549 GR cells increased dramatically to 3.8 folds of normal A549 cells (Fig. 2B, P<.001). This phenomenon provoked our further investigation of the role of MALAT1 on gefitinib resistance. Our results showed that the transfection of shMALAT1 in A549 GR cells decreased the percentage of survival cells at different concentrations of gefitinib including 0.005, 0.01, 0.05, 0.1, 0.5 and 1mM, when compared to the A549 cells transfected with control shRNA (Fig. 2C, P<.001). To validate the promotion of MALAT1 on gefitinib resistance, we overexpressed MALAT1 in normal A549 cells using MALAT1 expression plasmid (Fig. 2D), and tested the resistance of A549 cells against gefitinib at 0.005, 0.01, 0.05, 0.1, 0.5 and 1mM. Our results clearly showed that the A549 cells with MALAT1 overexpression demonstrated significantly higher gefitinib resistance than the cells transfected with normal plasmid (Fig. 2E, P<.001). Similarly, higher gefitinib resistance could be observed in the HC1299 cells with MALAT1 overexpression when compared to the cells transfected with normal plasmid (Fig. 2F, P<.001).
LncRNA MALAT1 promotes gefitinib resistance in lung cancer cells. (A) The gefitinib resistant A549 GR cells was made resistant to gefitinib by growing it in increasing concentrations of gefitinib. (B) The mRNA levels of MALAT1 in A549 GR or parental cells were determined by qPCR. (C) Cell viability of A549 GR cells transfected with or without MALAT1 shRNA treated with different concentration of gefitinib was determined by MTT assay. (D) The mRNA levels of MALAT1 in A549 cells transfected with MALAT1 expression plasmid were determined by qPCR. (E) Cell viability of A549 cells transfected with or without MALAT1 expression plasmid treated with different concentration of gefitinib was determined by MTT assay. (F) Cell viability of HCC1299 cells transfected with or without MALAT1 expression plasmid treated with different concentration of gefitinib was determined by MTT assay. ** P < .01; *** P < .001.
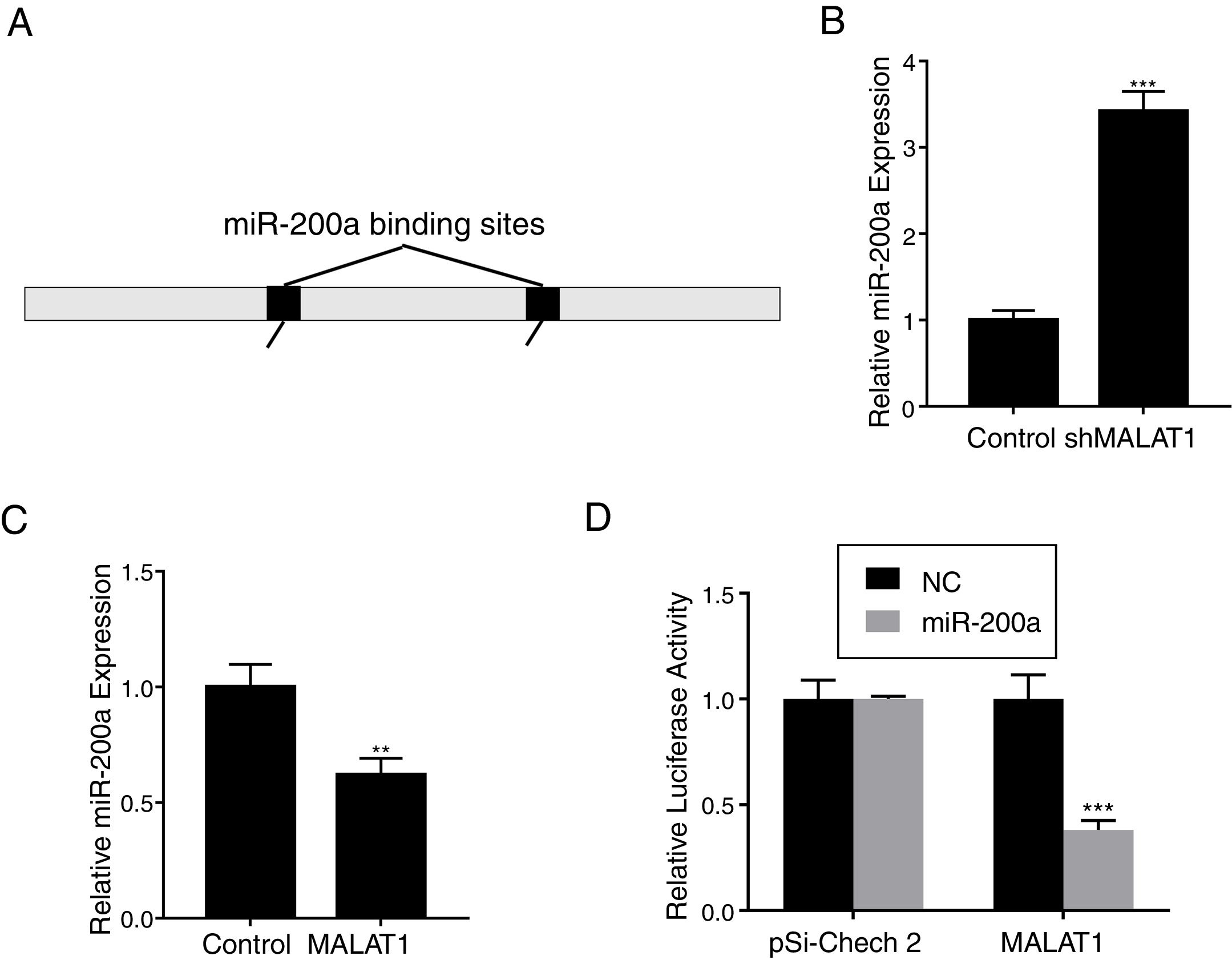
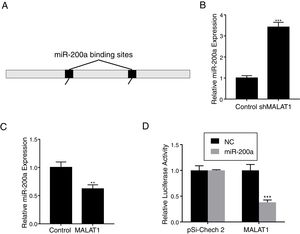
To explore the mechanism underlying the effects of LncRNA MALAT1 on cancer cells, bioinformatics analysis was conducted to identity the potential target of MALAT1. Using starBase v 2.0, we found that there are two miR-200a binding sites on the MALAT1 gene, which locate at positions 3444–3466, and 5435–5458 respectively (Fig. 3A). RT-PCR showed that the relative miR-200a expression in the A549 cells transfected with shMALAT1 was significantly elevated when compared to the cells with control shRNA (Fig. 3B, P<.001). As predicted, the A549 cells with MALAT1 expression had lower relative miR-200a expression (Fig. 3C, P<.01). To confirm the binding between miR-200a and LncRNA MALAT1, the luciferase reporter assay was performed using pSi-Check 2 luciferase reporter. In Fig. 3D, we could see the same level of fluorescence between the control and miR-200a in pSi-Check 2 vector. However, in the pSi-Check 2 vector with MALAT1 gene, miR-200a significantly inhibited the fluorescence (P<.001), suggesting the direct binding between MALAT1 and miR-200a.
LncRNA MALAT1 functions as a miR-200a sponge in lung cancer. (A) Schematic miR-200a putative target sites in MALAT1. (B) The mRNA levels of miR-200a in A549 cells transfected with MALAT1 shRNA were determined by qPCR. (C) The mRNA levels of miR-200a in A549 cells transfected with MALAT1 expression plasmid were determined by qPCR. (D) Luciferase reporter assay of pSi-check2 plasmid contains MALAT1 and transfected with miR-200a or NC. ** P < .01; *** P < .001.
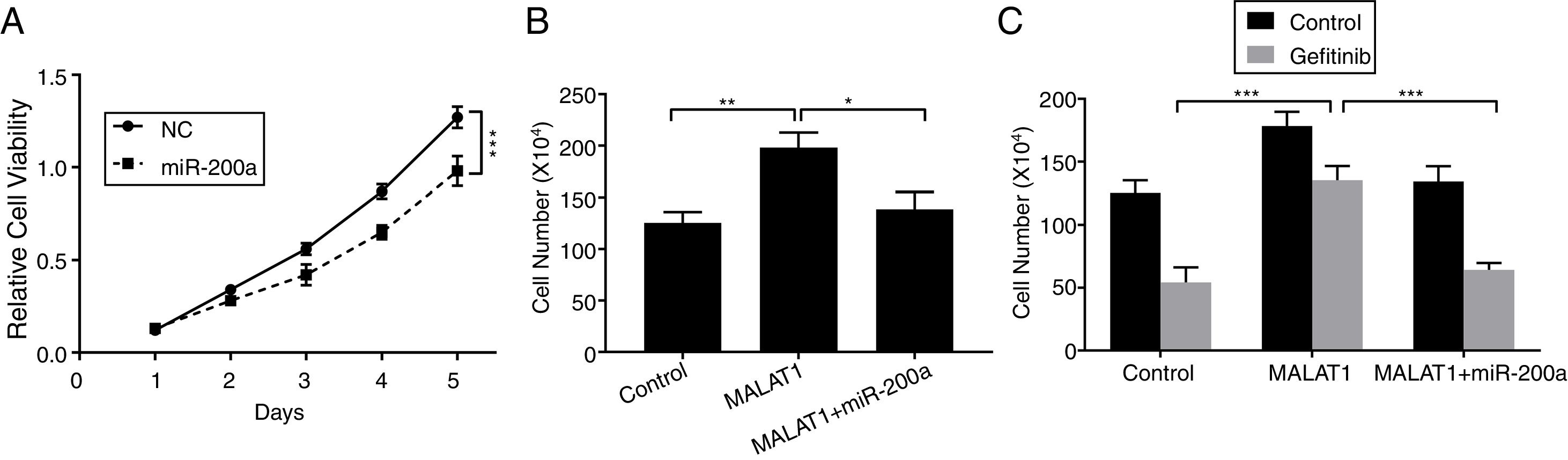
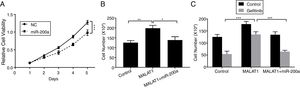
To this end, we observed the promotion of MALAT1 on tumor cell proliferation, and the interaction between MALAT1 and miR-200a. To determine whether the tumor promotion of MALAT1 is related to miR-200a, we transfected the A549 cells with miR-200a. The results showed that the relative cell viability of cells transfected with miR-200a was obviously decreased than the one of control cells (Fig. 4A, P<.001). Further, from Fig. 4B, cells with the overexpression of MALAT1 showed higher cell numbers than normal cells (P<.01), while co-transfection of miR-200a decreased the cell numbers (P<.05). Moreover, the MALAT1 overexpression demolished the damage of gefitinib on the A549 cells (Fig. 4C, P<.001), which was consistent with the promoted gefitinib resistance of A549 cells. Interestingly, cells with co-transfection of miR-200a showed decreased cell number, indicating the impaired protective function of MALAT1on gefitinib.
LncRNA MALAT1 sponging miR-200a to promote lung cancer proliferation and gefitinib resistance. (A) The proliferation of A549 cells transfected with miR-200a mimics or negative control (NC) was determined by MTT assay. (B) The proliferation of A549 cells transfected with miR-200a mimics and/or MALAT1 expression plasmid was determined by cell counts assay. (C) The proliferation of A549 cells transfected with miR-200a mimics and/or MALAT1 expression plasmid treated with or without 0.05μM gefitinib was determined by cell counts assay. * P < .05; ** P < .01; *** P < .001.
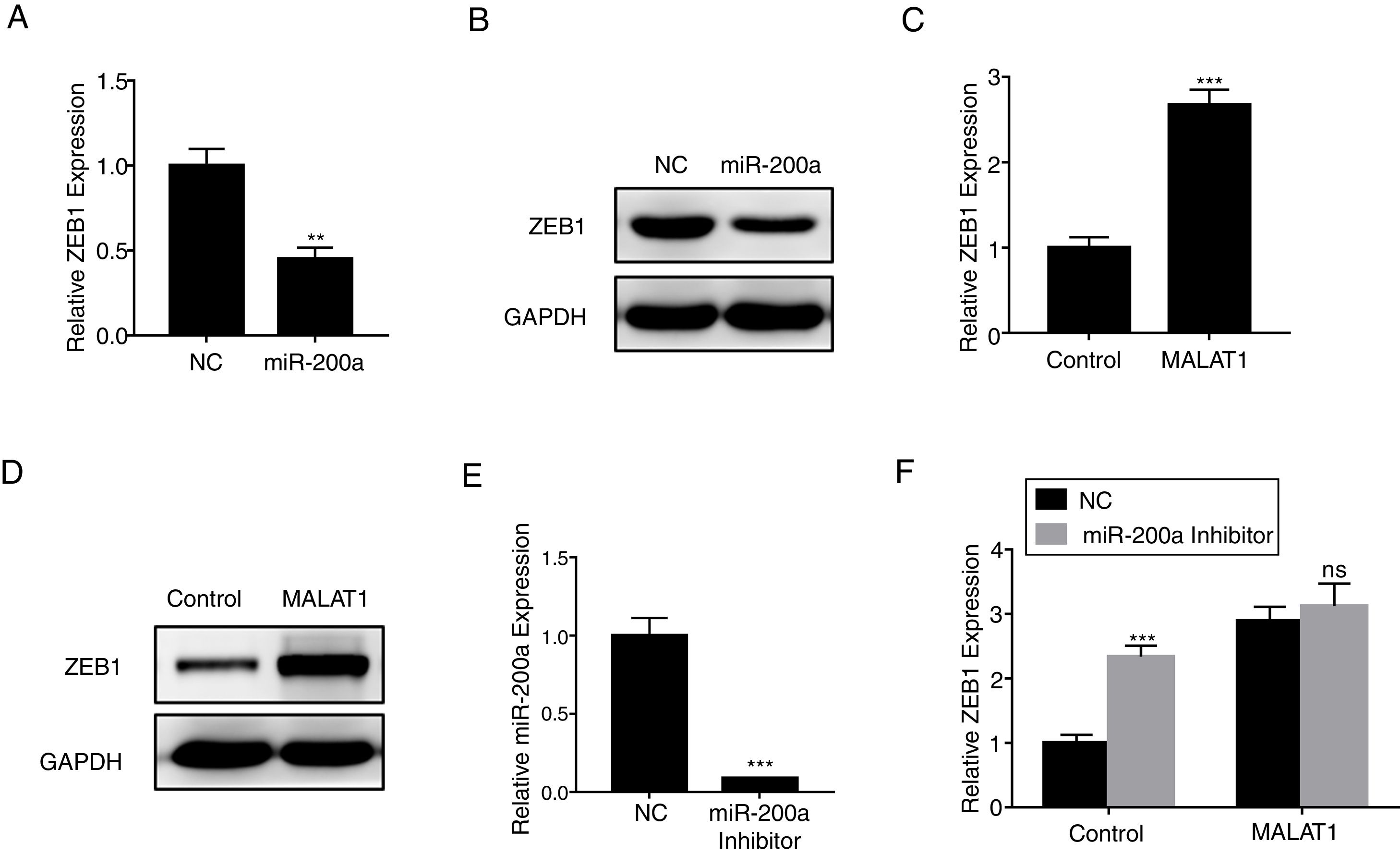
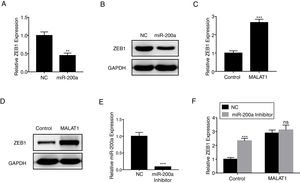
ZEB1 has been known as the down steam target of miR-200a,16 herein we confirmed that miR-200a downregulated the relative expression of ZEB1 in A549 cells at mRNA level (Fig. 5A, P<.01), and protein level (Fig. 5B). RT-PCR results also showed that the overexpression of MALAT1 in A549 cells upregulated the relative ZEB1 expression at both mRNA level (Fig. 5C, P<.001), and protein level (Fig. 5D). Furthermore, a miR-200a inhibitor was used in the assay to inhibit the expression of miR-200a (Fig. 5E). When we applied the same miR-200a inhibitor to A549 cells, we found the expression of ZEB1 was significantly upregulated (P<.001). In contrast, the application of miR-200a inhibitor did not present significant effects on the ZEB1 expression in the A549 cells with MALAT1 overexpression (Fig. 5F).
LncRNA MALAT1 promotes ZEB1 expression. (A) The expression of ZEB1 in A549 cells transfected with miR-200a was determined by qPCR. (B) The expression of ZEB1 in A549 cells transfected with miR-200a was determined by western blot. (C) The expression of ZEB1 in A549 cells transfected with MALAT1 expression plasmid was determined by qPCR. (D) The expression of ZEB1 in A549 cells transfected with MALAT1 expression plasmid was determined by western blot. (E) The expression of miR-200a in A549 cells transfected with miR-200a Inhibitor was determined by qPCR. (F) The expression of ZEB1 in A549 cells transfected with MALAT1 expression plasmid and/or miR-200a Inhibitor was determined by qPCR. ns, not significant. ** P < .01; *** P < .001.
The eukaryotic genome holds on to a large quantity of noncoding RNAs, which compose small and long noncoding RNAs (lncRNAs), the latter of which have recently been discovered as important molecules in a great variety of cellular processes.17 Various lncRNAs display abnormal expression in human cancer specimens and are involved in a number of oncogenic or tumor suppressor pathways.18 MALAT1 is one of the first identified cancer-associated lncRNAs, which was originally reported as a prognostic marker for metastasis and survival of NSCLC patients.19 In the current study, our group aimed to explore the possible mechanisms and potential therapeutic roles of lncRNA MALAT1 in lung cancer. The results demonstrated that knockdown of MALAT1 by shMALAT1significantly inhibited the cell viabilities and cell proliferation in tumor cells A549 and HCC1299, indicating the promoting role of lncRNA MALAT1 in lung cancer cell proliferation. In addition, we observed that lncRNA MALAT1 promoted gefitinib resistance in A549 cells. Further mechanism investigation revealed that MALAT1 functioned as a miR-200a sponge, which enhanced ZEB1 expression in A549 cells, ultimately promoting the cell proliferation and gefitinib resistance.
MicroRNAs are a class of small single-stranded RNAs, which are approximately 20–24 nucleotides long.20 Recent studies have demonstrated that LncRNA MALAT1 functioned as miRNA sponges. For instance, LncRNA MALAT1 was revealed to promote colorectal cancer cell proliferation, invasion and migration by downregulating miR-145,21 and to enhance the docetaxel chemoresistance of prostate cancer cells targeting miR-145-mediated AKAP12.22 In addition, MALAT1 silencing suppressed the progression of prostate cancer by miR-1 upregulation.23 MALAT1 facilitates cell proliferation and activates autophagy by sponging miR-101 in glioma.24 MALAT1/miR-124/Capn4 axis modulates proliferation and invasion of nasopharyngeal carcinoma cells.25 Researchers have revealed that MALAT1 regulated miR-145 in gastric cancer by acting as a competing endogenous RNA.26 As for miR-200, miR-200c/MALAT1 axis could serve as therapeutic target in endometrioid endometrial carcinoma.27 In line with this, we found that MALAT1 binds miR-200a using bioinformatics analysis, and the interaction was validated by luciferase reporter assay. In addition, knockdown of MALAT1 via shMALAT1 largely promoted relative miR-200a expression, while overexpression of MALAT1strikingly suppressed the miR-200a expression.
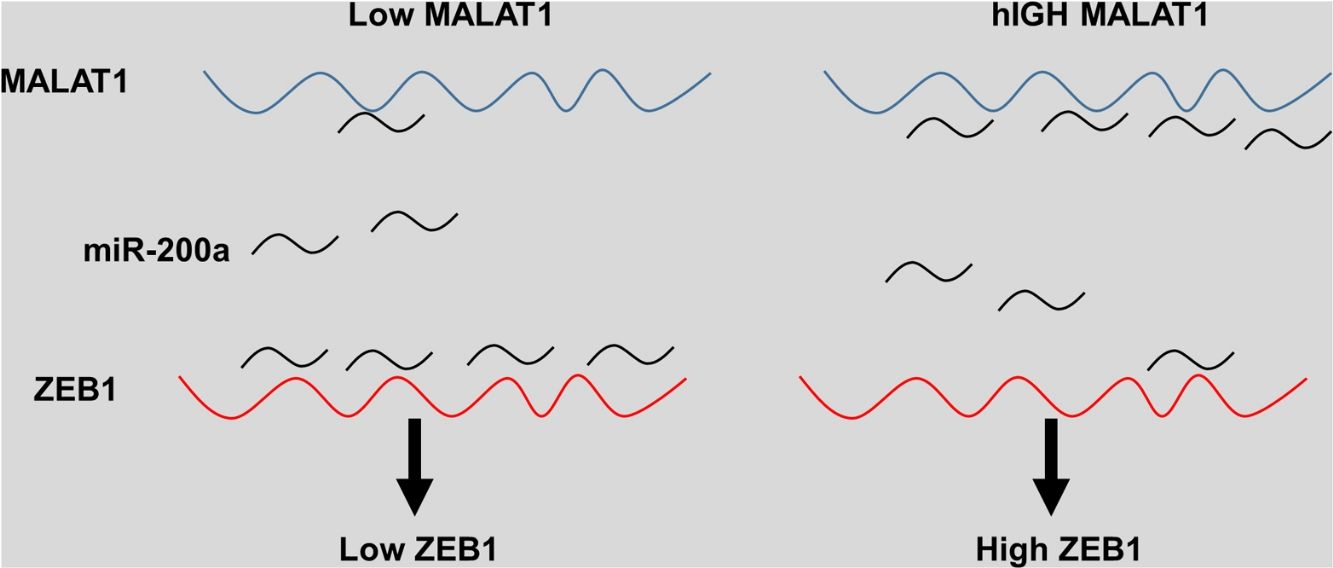
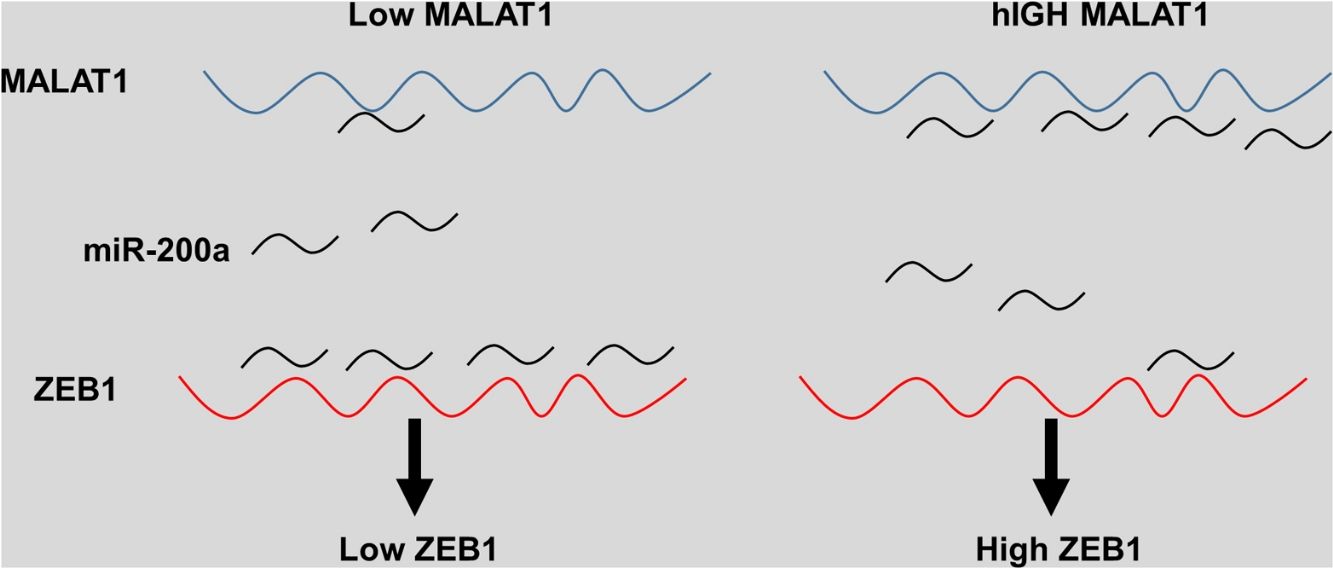
The miRNA-200 family consists five members including miR-200a, miR-141, miR-200b, miR-200c and miR-429.28 Previous studies have shown that the expression of miR-200 could be inhibited by several transcription factors. Members of MiR-200 family inhibit epithelial-to-mesenchymal transition (EMT) by targeting zinc-finger E-box-binding homeobox factor 1 (ZEB1) and 2 (ZEB2) directly.29 Intriguingly, ZEB1 and ZEB2 suppress the activity of miR-200 promoter through binding to the E-box elements, indicting the existence of a feedback loop between ZEB and miR-200.30 Based on the results that knockdown MALAT-1 can inhibit lung cancer cells proliferation, we hypothesized that these effects may be modulated by regulating ZEB genes. As predicted, we noticed that the relative ZEB1 expression of lung cancer cells transfected with miR-200a was downregulated. Moreover, lung cancer cells added with miR-200a showed significantly upregulated ZEB1 expression. Intriguingly, we observed upregulation of relative ZEB1 expression in lung cancer cells with MALAT1 overexpression, but this higher expression was not inhibited by miR-200a inhibitor. We speculated that overexpression of MALAT1forfeited the promotion of miR-200a completely, thus the addition miR-200a could not make significant difference on the ZEB1 expression, indicating the existence of MALAT1/miR-200a axis.
LncRNAs have been widely reported to be associated with gefitinib resistance in tumor cells. By analyzing human lung cancer cells resistant or sensitive to EGFR-TKI through lncRNA microarray, previous research showed that the expression of a large number of lncRNAs were different in gefitinib-resistant cells.31 Some other examples included the contribution of MIR31HG lncRNAs to gefitinib resistance in PC9 cells,32 and the role of GAS5 LncRNA in the development of gefitinib resistance in A549 cells.32 LncRNA BC087858 could induce EGFR-TKIs resistance and promote cells’ invasion via the activation of MEK/ERK and PI3K/AKT pathways in NSCLC.33 In our study we demonstrated the promotion of LncRNA MALAT1 on the gefitinib resistance in A549 cells. It has been reported that miR-200a expression is able to render the sensitivity of lung cancer cells to gefitinib treatment.34 Previous study also showed that the miR-200/ZEB axis could serve as predictive biomarkers for NSCLC cells sensitivity to nintedanib, which is a multi-targeted angiokinase inhibitor.35 Further, another study showed that polyphyllin exhibited antitumor activity against gefitinib-resistant NSCLC via mechanism associated with the downregulation of MALAT1.15 In this study, we accordingly investigated the effects of LncRNA and miR-200a sponge on the gefitinib resistance. Our results showed that the promotion of MALAT1 on gefitinib resistance was related to miR200-a, since the transfection of miR200-a neutralized the protection of MALAT1 overexpression in the gefitinib damage.
To summarize, our study showed that MALAT1 promoted the proliferation of lung cancer cells by interacting with miR-200a. Furthermore, we applied miR-200a inhibitor to A549 cells, and the results showed the potential therapeutic effects of miR-200a in lung cancer cell proliferation. In addition, the MALAT1/miR-200a sponge acts by regulating expression of ZEB1 in the cells. However, our study was limited by the sole in vitro system used. The A549 and HCC 1299 human lung adenocarcinoma cell lines might not stand for the real pathological condition occurring in the tumors of patients. We will further validate the discovery in clinical patient samples. Taken together, these findings indicated that the MALAT1/miR-200a sponge revealed the possible mechanism underlying the gefitinib resistance of NSCLC, and may represent a novel potential therapeutic target for lung cancer treatment.
ConclusionOur study showed that MALAT1 promoted the proliferation and gefitinib resistance of lung cancer cells by sponging miR-200a, which regulates expression of ZEB1 in the A549 cells. This MALAT1/miR-200a axis could serve as new therapeutic target for lung cancer treatment.
FundingThis study was supported by National Natural Science Foundation of China (#21876205 and 81573026).
Conflict of InterestsThe authors declare no conflict of interests.