Hereditary alpha-1-antitrypsin (α1-AT) deficiency predisposes to pulmonary emphysema. The objective of this study is to demonstrate the limitations of some laboratory methods used in the study of the deficiency, and which may produce errors in interpretation and detection of uncommon alleles.
Two clinical cases are described: the index patient, who had pulmonary emphysema with α1-AT levels less than 12mg/dL, was erroneously classified as a homozygote of the normal allelic variant PI MM using a rapid genotype method; the mother of the patient, asymptomatic, with low levels (60mg/dL), was also classified as PI MM.
The gene sequencing classified the index patient as a carrier of the PI Clayton null allele and PI Mmalton deficient. The mother was a PI Clayton/PI heterozygote carrier.
These results highlight the difficulties in diagnosing the deficiency, as the well as the need to reach a consensus on methods for this study.
El déficit hereditario de alfa 1-antitripsina (α1-AT) predispone al desarrollo de enfisema pulmonar. El objetivo de este trabajo es evidenciar las limitaciones de algunos métodos de laboratorio utilizados en el estudio del déficit, cuya interpretación puede inducir a error en la detección de alelos poco frecuentes.
Se describen dos casos clínicos: la paciente índice, que presentaba enfisema pulmonar con niveles de α1-AT inferiores a 12mg/dL, catalogada erróneamente como homocigota de la variante alélica normal PI MM mediante un método de genotipificado rápido; la madre de la paciente, asintomática, con niveles bajos (60mg/dL), catalogada también como PI MM.
La secuenciación del gen catalogó a la paciente índice como portadora del alelo nulo PI Clayton y del deficitario PI Mmalton. La madre resultó portadora heterocigota PI Clayton/PI M.
Los resultados ponen de relieve las dificultades de diagnóstico del déficit así como la necesidad de consensuar métodos para este estudio.
Alpha-1 antitrypsin deficiency (α1-AT) is a hereditary disorder associated with pulmonary emphysema in adults.1 The main physiological function of α1-AT, also known as a proteinase inhibitor (PI), is to protect the lung tissue from the proteolytic enzymes.2 The alleles most commonly associated with normal α1-AT levels are in general PI M. However, in addition to the normal PI alleles there are also uncommon PI M, which are deficient.3,4 The alleles most commonly associated with low levels of α1-AT are PI S and PI Z, and the majority of individuals with severe deficiency are PI Z homozygous.5,6 The deficiency can also be due to other alleles, either dysfunctional or null.7,8
The diagnosis of deficiency involves the quantification of α1-AT and the later phenotyping or genotyping when the concentration is low.9 However, some fast genotyping methods can easily led to erroneous diagnoses, as some uncommon alleles, and the null ones in particular, are not detected. This is the case of the so-called allele-specific techniques that only detect the PI S and PI Z alleles.10–13
This clinical observation reflects the errors in the diagnosis of two cases with α1-AT deficit, heterozygous carriers of PI Clayton null allele, who had been considered normal PI MM homozygous by means of a method of allele-specific hybridization that had been inadequately interpreted.
Clinical ObservationWe included for study two members of the same family. The index case is a 35-year-old woman who is an ex-smoker. She presented symptoms of bronchial hyperreactivity associated with infectious processes of the lower airways. The determination of the levels of α1-AT was clearly deficient: 12mg/dL (normal range: 110–190mg/dL).
Chest CT revealed signs of diffuse panacinar emphysema with a greater affectation in the lower lobes and signs of paraseptal emphysema in both apex and upper lobes. The functional tests with bronchodilator treatment were: FVC 3.39L (93% pred), FEV1 1.34 L (42% pred), FEV1/FVC 40, TLC 7.34 (138% pred), RV 3.95 L (247% pred), DLCO 5.9 (64% pred), DLCO/AV 1.11 (53% pred).
Due to the low concentration of α-1AT, the phenotype could not be determined. For this reason, we opted for determining the genotype by means of a method of allele-specific hybridization that only detects the PI S and PI Z variants. In this analysis, the patient was catalogued as a non-carrier of these alleles, and by exclusion she was erroneously reported as a carrier of the most common normal variant of α1-AT, PI MM. The fast clinical progression and the low level of α1-AT led to the establishment of substitutive treatment (180mg/kg/21 days of α1-AT) while the disagreement between the levels and the assigned genotype was investigated by sequencing.
The mother of the patient, who was 71 years old and asymptomatic from a respiratory standpoint, presented an α1-AT of 60mg/dL and had also been catalogued, by the same allele-specific hybridization method, as a homozygous PI MM carrier. This incoherence also lead to the study of the genotype.
In both cases, the determination of the α1-AT genotype was carried out in DNA isolated from total blood by means of a procedure of amplification and secuencing.14
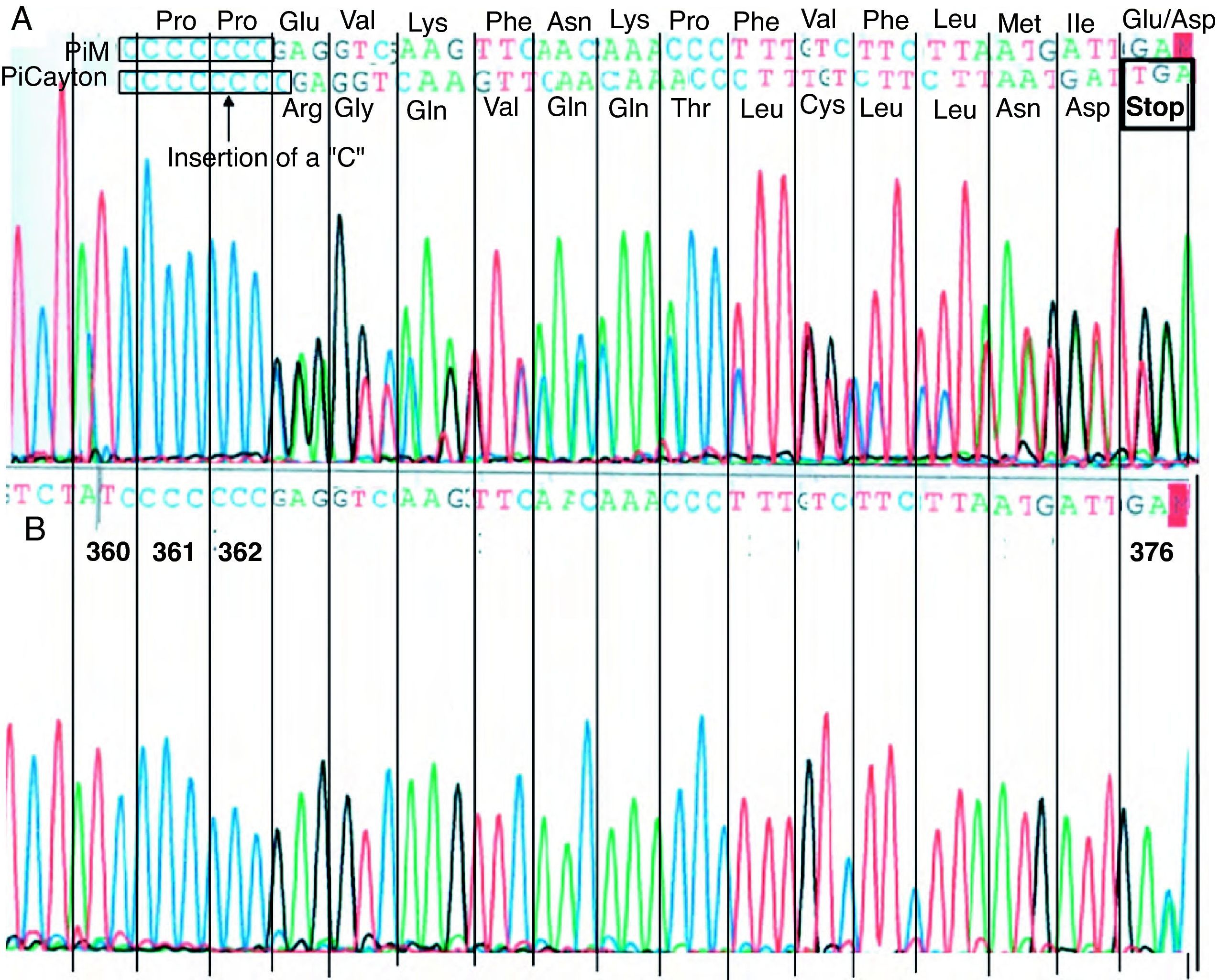
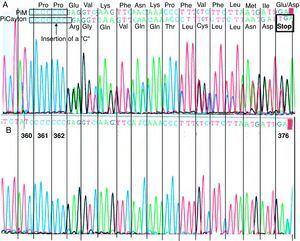
The analysis of the DNA sequencing showed that the index case presented two different mutations corresponding with the helical variants PI Clayton and PI Mmalton: PI Clayton is a null variant of α1-AT characterized by the insertion of a cytosine in the V exon of the gene.15 As a consequence of this insertion to the protein, it lacks 19 amino acids. Fig. 1 shows a fragment of the V exon in which one can observe the mutation corresponding to the PI Clayton variant. PI Mmalton is an uncommon deficient variant characterized by the deletion of three pairs of bases (TTC) in exon II.16 The PI Mmalton variant is associated with a concentration of α1-AT similar to that observed in a PI ZZ individual (de 15–35mg/dL, approximately). The patient was considered heterozygous for the two deficient variants.
Direct sequencing of a V exon fragment from the α1-AT gene in the region corresponding to the allele PI Clayton. PI Clayton is a null variant of α1-AT characterized by the insertion of a cytosine in the V exon of the normal M1 allelic sequence (Val213) that usually has seven cytosines (C) that codify the amino acids 360–362. (A) Sequencing corresponding with the index case, heterozygous for the PI Clayton variant. In the upper part of the figure, the two overlapping sequences are indicated: PI M (non-mutated sequence) with seven cytosines and PI Clayton with eight cytosines. (B) Reference sequencing of the same fragment from a patient without the mutation.
The family analysis showed that the mother was heterozygous for the PI Clayton and the normal PI M allele.
DiscussionThe null alleles of α1-AT are defined as genes in which, due to a mutation in DNA sequencing, the protein is not expressed.7,17,18 They are extremely rare alleles, and 16 types have been reported.8 Our group, which, as the laboratory for the Spanish α1-AT Deficiency Registry, has been studying this disease since 1993, had only detected one case that of a COPD patient carrier for null allele PI Bellingham. The clinical importance of the null alleles becomes more evident when they are inherited, either with another null or with another deficient allele.
In this clinical observation, we describe the case of a patient with a history of severe pulmonary emphysema, with substitutive treatment with intravenous α1-AT, heterozygous carrier of the null allele PI Clayton. From the available data, this is the first known case of a patient who is a heterozygous carrier of the null allele PI Clayton together with the uncommon deficient allele PI Mmalton associated with the intracellular degradation of α1-AT.16 The mother of the patient, who was asymptomatic, was heterozygous for the allele PI Clayton and the normal PI M. The two patients had been catalogued erroneously as homozygous carriers of the normal allele PI M. The null PI Clayton allele has previously been detected, associated with pulmonary emphysema, also in a state of heterozygosis with uncommon deficient variants of α1-AT, in the United States and in Japan.19
The clinical cases analyzed demonstrate some of the potential problems associated with the diagnosis of α1-AT deficiency. Due to the fact that the index case had very low levels of α1-AT in serum, the α1-AT phenotype could not be determined. As a consequence, we opted for the analysis of the genotype using an allele-specific hybridization technique that only detects the PI S and PI Z variants. The patient was catalogued as non-carrier of these two alleles and by exclusion as carrier of the normal PI MM alleles, considering that if in the general population the most common alleles are PI M, PI S and PI Z, the combination PI MM would be most likely. A similar error occurred in the genotype of the mother. The allele-specific techniques are quick, easily automatable and low-cost, but give partial genotyping, identifying only some alleles.11 They simply report the presence or absence of the alleles studied. Therefore, it should never be deduced (although it is quite likely) that if these alleles are not identified we are dealing with a normal allele. These methods are reliable, however, as the errors are mainly related with the incorrect interpretation of the results.
With the aim of establishing effective programs for detecting the greatest number of patients affected by α1-AT deficiency, a series of guidelines have been published.9,20 The determination of α1-AT levels and the phenotype in serum are the recommended laboratory analyses. For each phenotype, there is a specific corresponding range of α1-AT values. In the cases in which there is lack of agreement between the levels and the phenotype, the molecular characterization of the gene should be done using sequencing. It must be emphasized that the main application of allele-specific detection techniques is the screening of the PI S and PI Z alleles.
It is therefore important for each laboratory to report the method used to study the deficiency and its limitations, if any, as well as the reference values that correspond to each phenotype or genotype. Taking into account the fact that the majority of hospitals have sequencing equipment available, this technique for the molecular study of the entire α1-AT gene should be advocated in cases in which the diagnosis poses doubts.
Please cite this article as: Rodríguez-Frías F, et al. Diagnóstico del déficit de alfa 1-antitripsina: limitaciones de las pruebas de laboratorio de diagnóstico rápido. Arch Bronconeumol. 2011;47:415–7.