Recently, lung transplantation from donation after circulatory death (DCD) has become a routine clinical practice to address the donor shortage.1,2 However, lungs stored in warm conditions undergo more severe damage than those stored in cold conditions, and ischemia reperfusion injury (IRI) following warm ischemia (WIRI) is the most problematic issue in lung transplantation from DCD compared to donation from brain death donors (DBD). Although WIRI has been recognized as an enhancement of IRI following cold ischemia,3 recent studies highlight notable mechanistic differences.4–6 Furthermore, a comprehensive RNA sequencing analysis of lungs from a mouse WIRI model recently identified numerous genes expressed earlier than the transcription factor EGR-1, which has been considered the master regulator for pulmonary IRI.7–11 However, no specific drug exists to manage WIRI, emphasizing the need for further investigation. The objective of this study was to clarify the molecular mechanism of WIRI and identify therapeutic targets.
In this study, we analyzed human peripheral lung tissue biopsies collected 30min after reperfusion. Five matched cases each of DCD and DBD lung transplantation were selected from a Spanish institutional database. The transplantations were performed between May 2020 and December 2020. As shown in Supplemental Table 1, donor factors were matched for gender, age, and cause of death, and recipient factors were matched for gender, age, diagnosis, operative procedure, and perioperative outcome. H&E staining revealed that nuclear fragmentation was more pronounced in DCD lungs than in DBD lungs, indicating more evident DNA damage in DCD lungs (Supplemental Fig. 1A). This finding was supported by a TUNEL assay, which showed significantly higher TUNEL-positive cells in DCD lungs (p=0.0159) (Supplemental Fig. 1C), suggesting differences in DNA damage status due to IRI between DCD and DBD lungs.
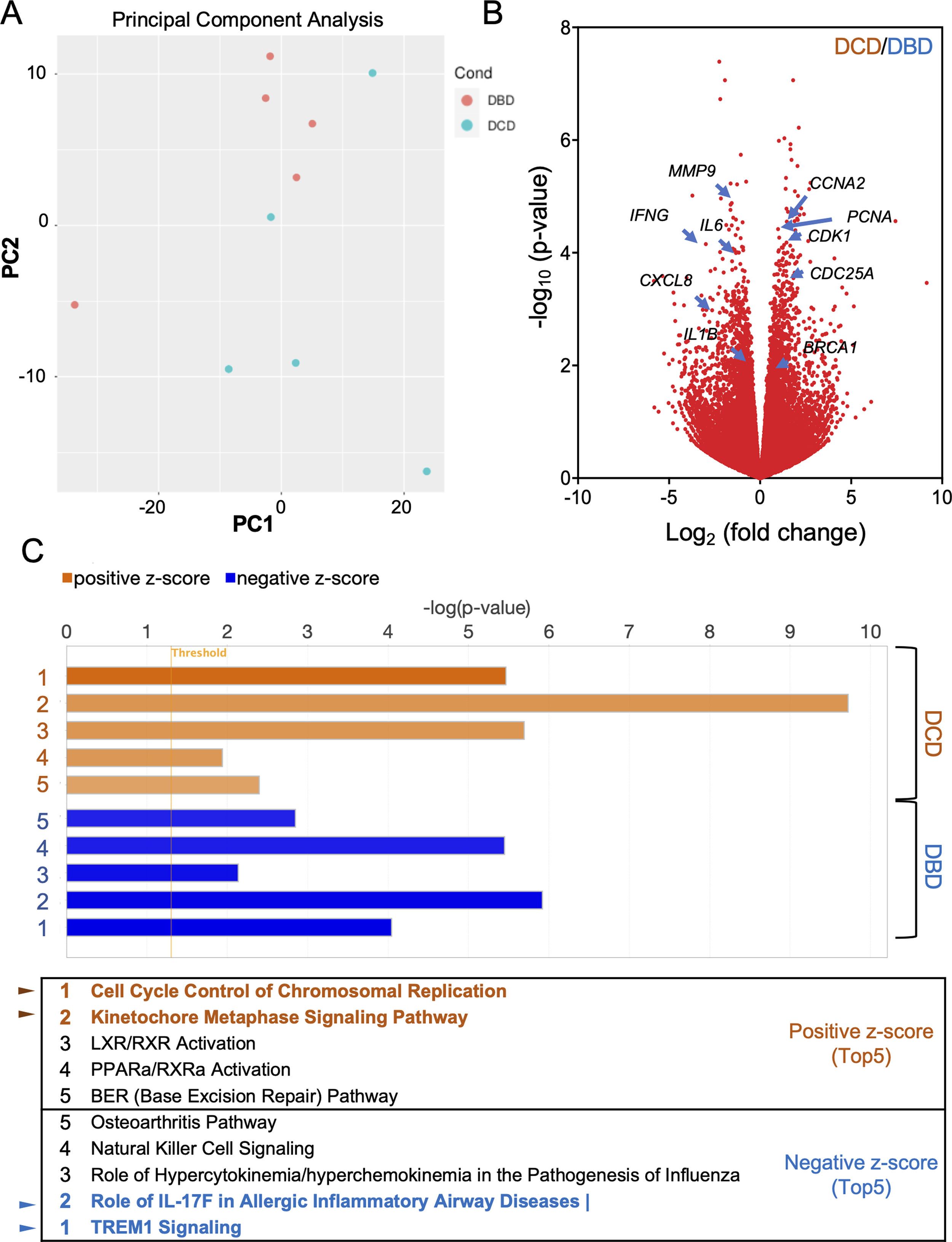
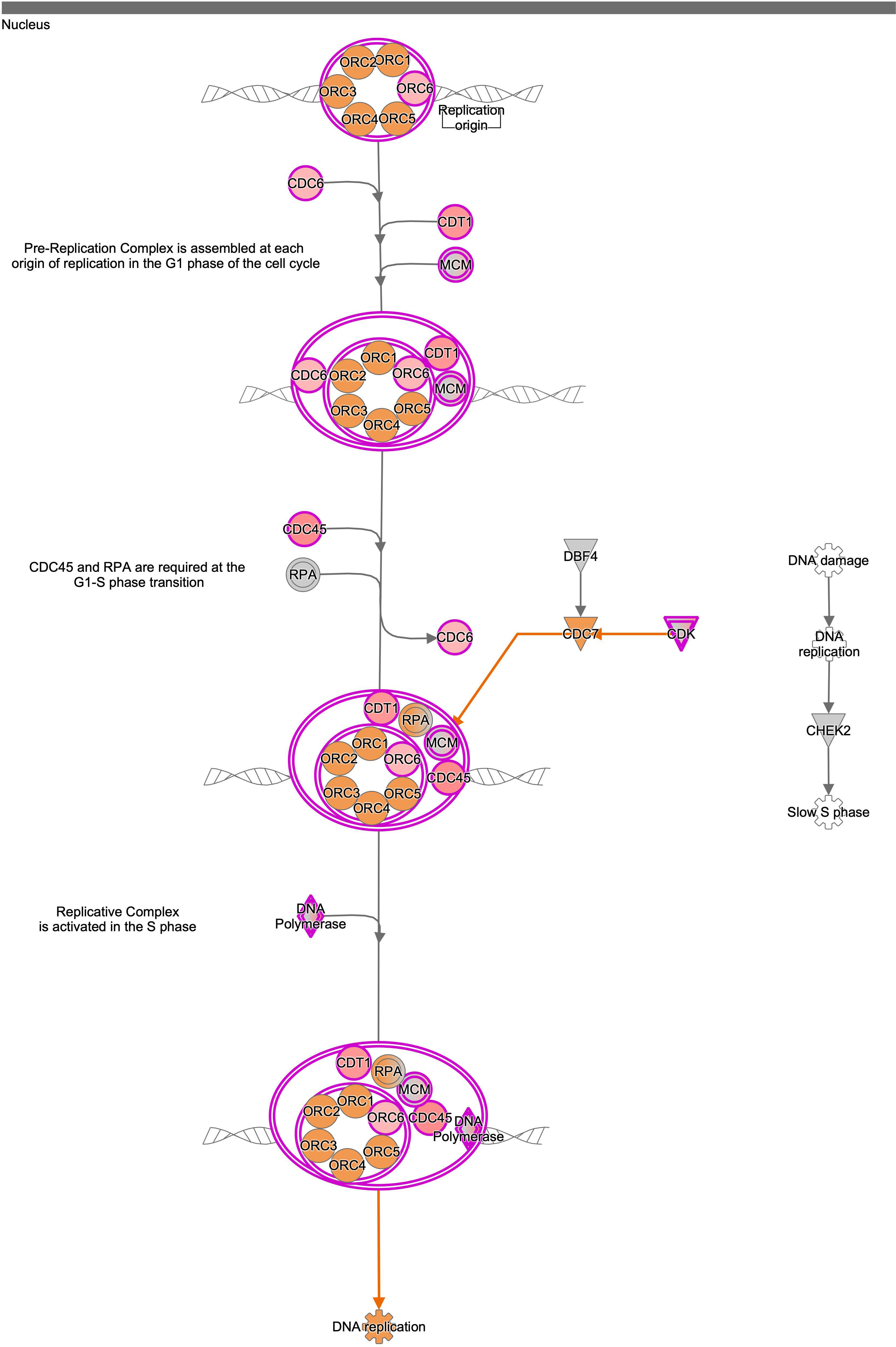
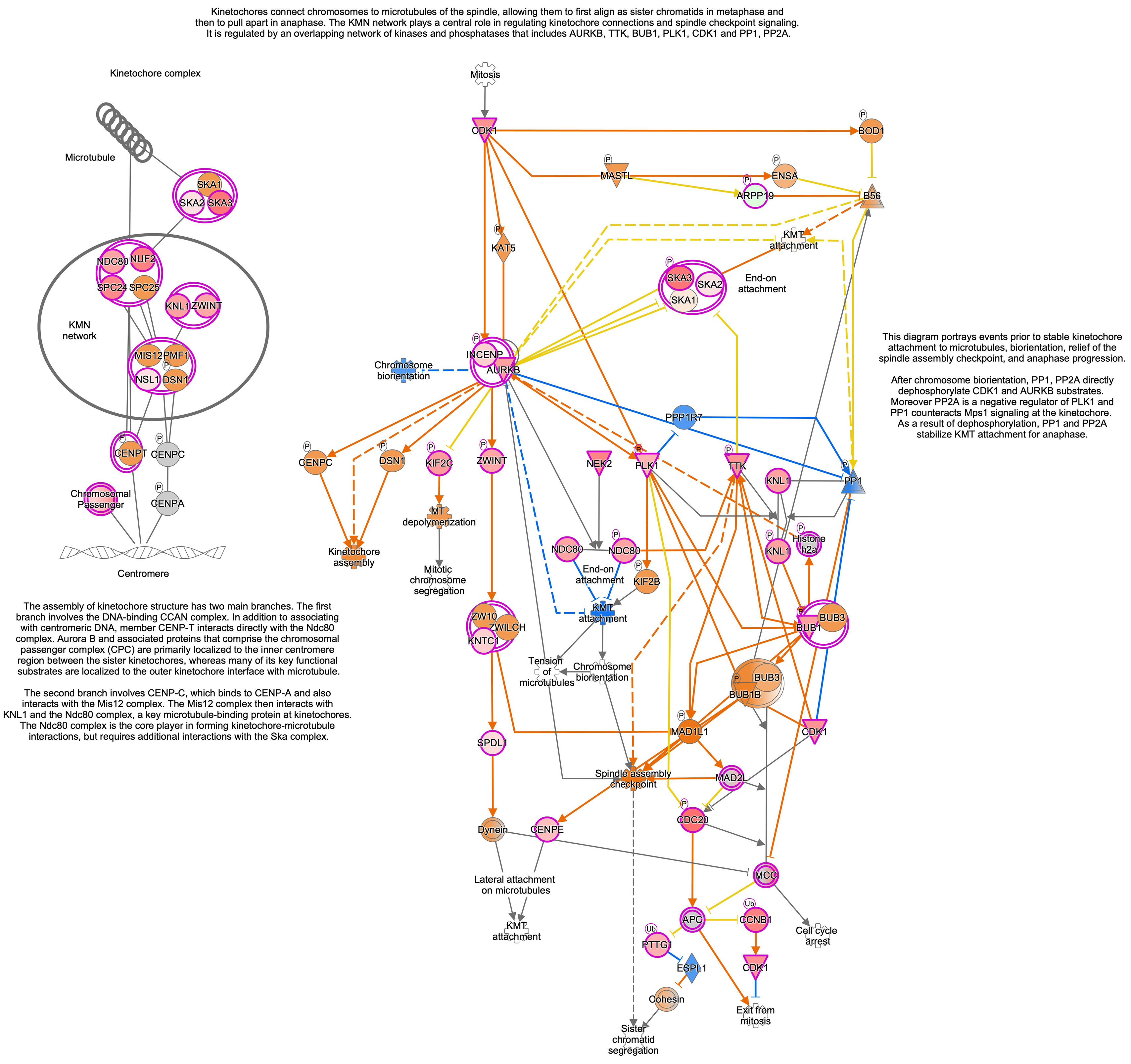
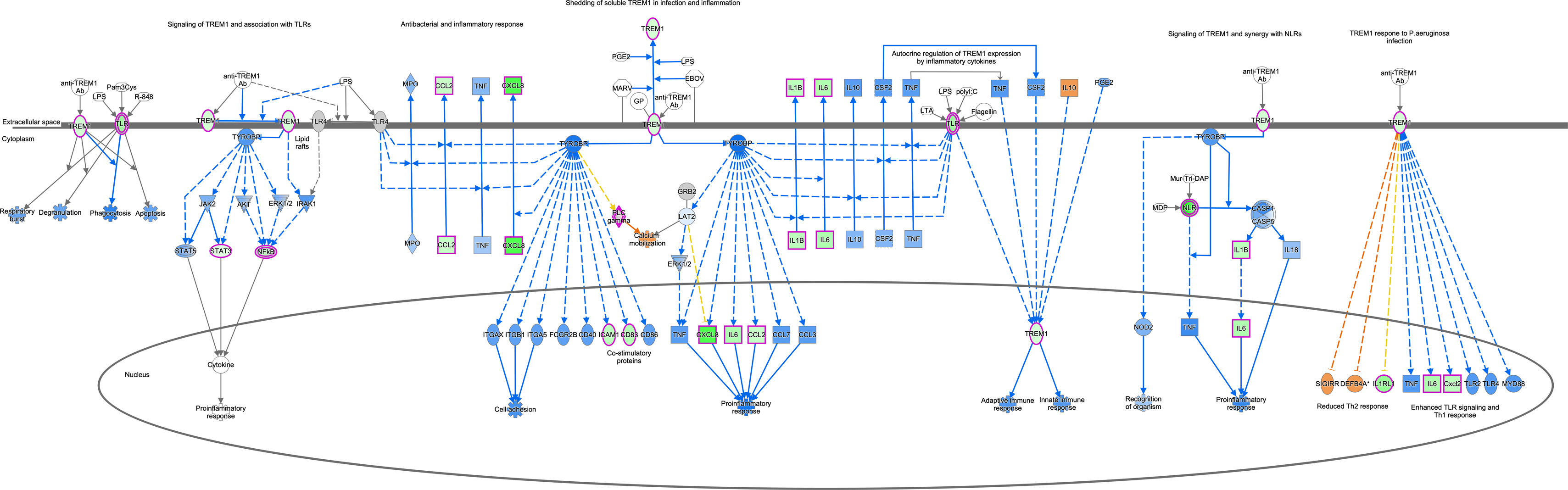
To understand the histological features of lung transplantations from DBD and DCD at molecular level, we conducted dynamic transcriptome analysis using RNA sequencing. Principal component analysis (PCA) demonstrated distinct RNA expression distributions between DBD and DCD lungs (Fig. 1A). Genes related to cell cycle and DNA damage and repair, such as PCNA, CCNA2, CDC25A, CDK1, and BRCA1, were more highly expressed in DCD lungs than in DBD lungs, while inflammation-related genes like MMP9, IFN-γ, IL-1β, IL-6, and CXCL-8 were more prominent in DBD lungs (Fig. 1B). Canonical pathway analysis further examined pathways associated with differentially expressed genes, revealing distinct pathways activated in DBD and DCD lungs (Fig. 1C). Specifically, genes associated with the Cell Cycle Control of Chromosomal Replication and Kinetochore Metaphase Signaling pathways were highly enriched in DCD lungs. Supplemental Figs. 2 and 3 show signal maps with upregulated genes in these pathways. Conversely, TREM1 signaling and the role of IL-17F in allergic inflammatory airway diseases were significantly upregulated in DBD lungs, as visualized in Supplemental Fig. 4. These findings underscore distinct gene expression profiles in DCD and DBD lungs, particularly in DNA damage response and inflammatory signaling, respectively.
Differences between lung transplantation from DBD and DCD by dynamic transcriptome analysis with RNA sequencing. (A) The distribution of RNA expressions in DBD and DCD lungs by principal component analysis. (B) Volcano plot represents genes highly expressed in each of DBD and DCD lungs. (C) Canonical pathway analysis represents pathways activated in DCD or DBD lungs. Orange: activation in DCD; blue: activation in DBD.
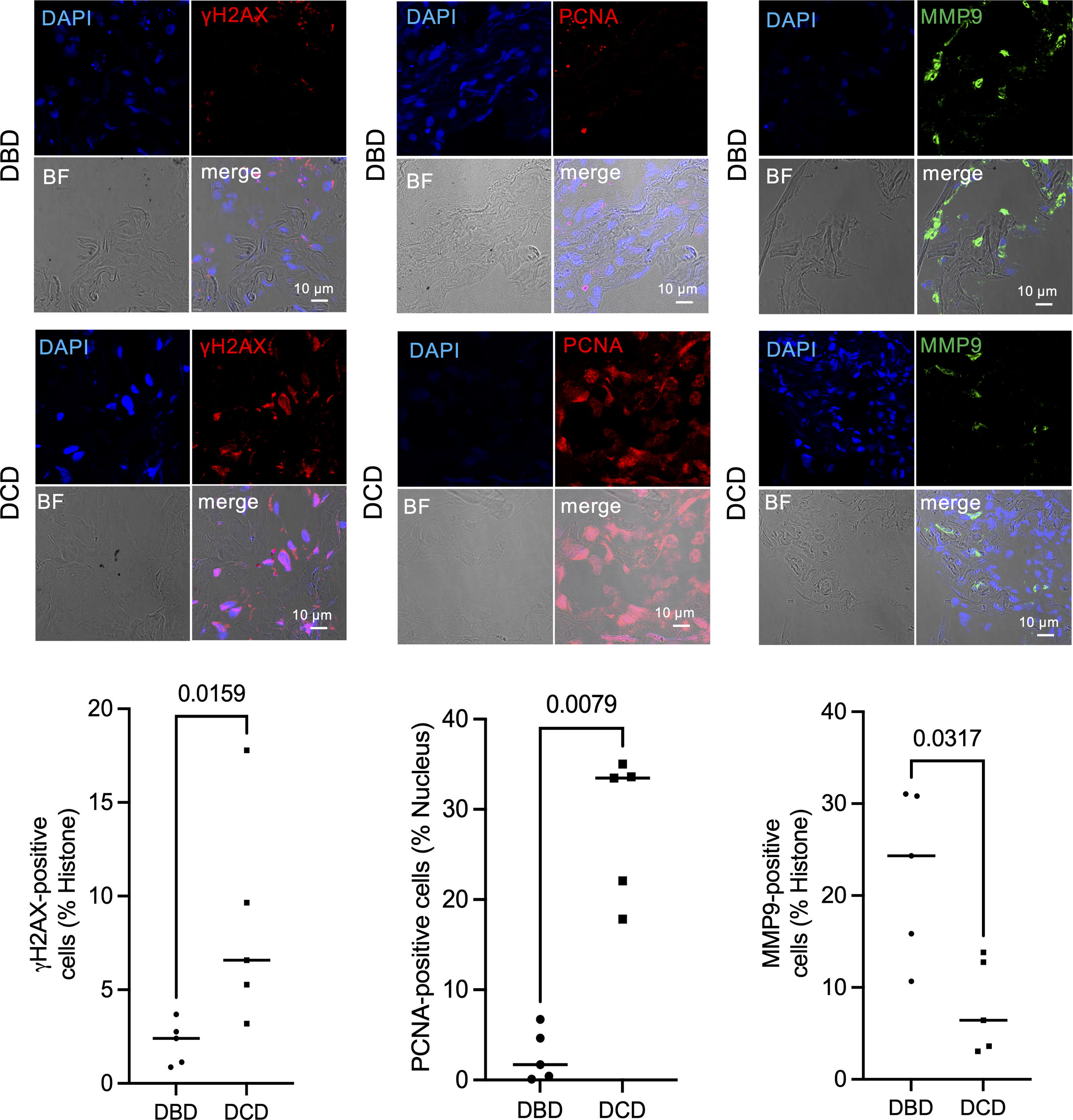
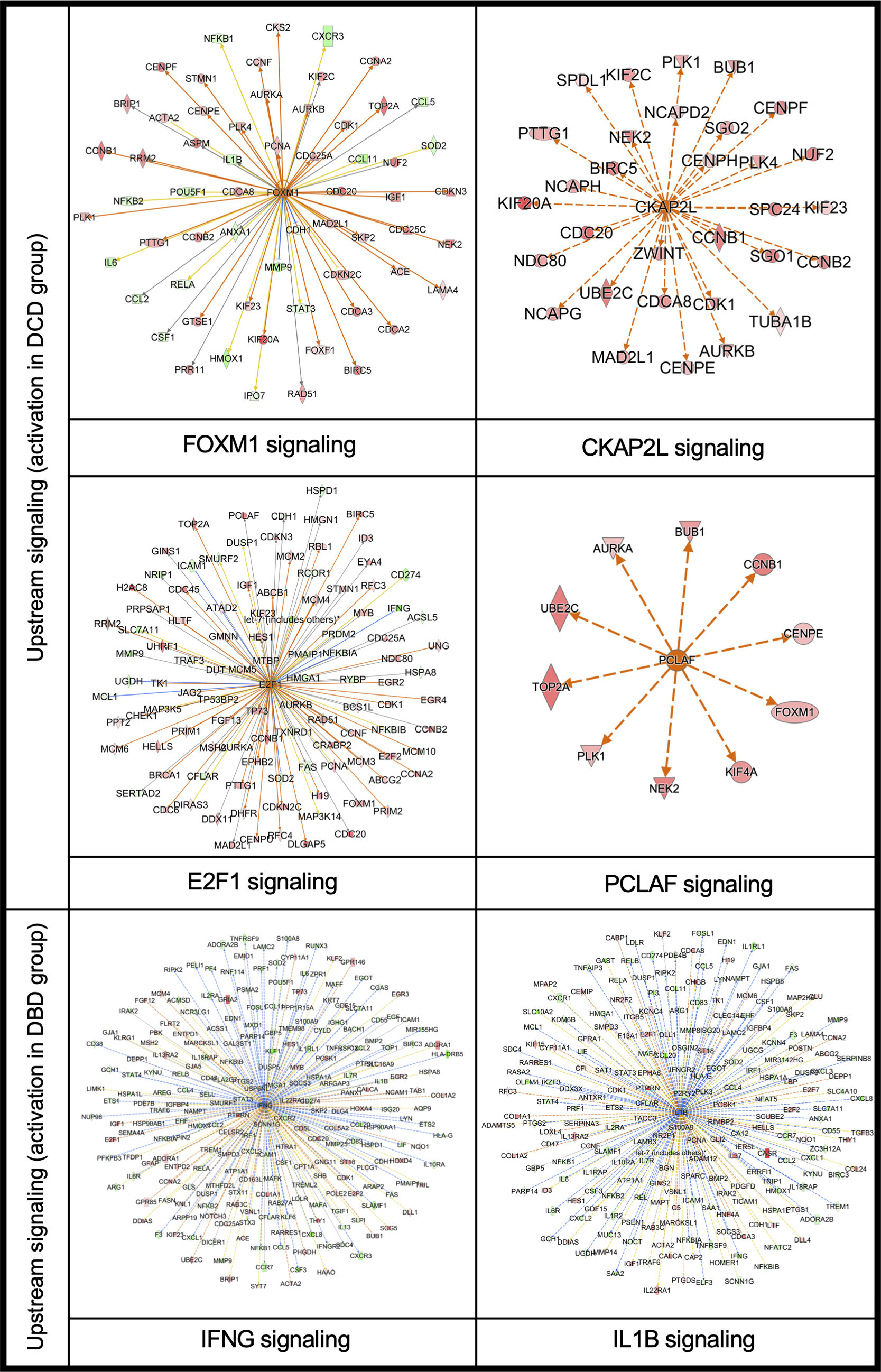
Further upstream analysis using IPA (Ingenuity® Pathway Analysis) clarified the clustering of RNA-sequencing data, identifying FOXM1, CKAP2L, E2F1, and PCLAF as upstream signals activated in DCD lungs (Supplemental Fig. 5). These signals are closely related to DNA damage and replication. Immunohistochemistry with anti-γH2AX antibody was used to quantify DNA damage at the protein level, with significantly more γH2AX-positive nuclei in DCD than in DBD lungs (Fig. 2). γH2AX-positive nuclei were significantly more in the DCD lungs than DBD lungs. γH2AX is a critical indicator of cellular response to DNA damage, particularly double-strand breaks,12–14 contributing to DNA repair, cell cycle regulation, and programmed cell death. While γH2AX is a sensitive DNA damage marker, no therapeutic agents targeting γH2AX have been reported, including in organ transplantation or cancer.
Immunofluorescence staining of γH2AX, PCNA, and MMP9 in DCD and DBD lungs. Representative immunofluorescence staining of γH2AX, PCNA, and MMP9 in DCD and DBD lungs, and number of γH2AX, PCNA, and MMP9 positive cells in DCD and DBD lungs. Data and bars are expressed as plots and medians (n=5).
To evaluate DNA replication levels in transplanted lungs, we quantified PCNA expression via immunofluorescence (Fig. 2). DCD lungs showed significantly more PCNA-positive cells than DBD lungs, indicating increased DNA replication. While cells routinely repair DNA damage caused by various stimuli, reactive oxygen species can induce DNA damage that activates poly ADP-ribose polymerase (PARP), which both repairs DNA and, under oxidative stress, promotes apoptosis and necrosis.15 Prior research by Hatachi et al. demonstrated that PARP inhibitors reduced inflammation and tissue damage in WIRI using a rat hilar clamp model.16
Despite limited studies on DNA repair targeting pulmonary IRI, Tan et al. reported that mitochondria-targeted administration of 8-oxoguanine DNA glycosylase-1 (OGG1), involved in DNA repair, suppressed IRI in a rat ex vivo model.17 They observed that OGG1 reduced mitochondrial DNA damage and decreased pro-inflammatory mitochondrial DNA fragments in lung perfusate. Ex vivo lung perfusion (EVLP) may offer a practical approach for DNA repair in DCD lungs. While stem cell, mesenchymal cell, and gene therapies are also potential treatments for DNA repair, no studies have demonstrated that these technologies improve transplanted lung function.
In DBD lungs, IFN-γ and IL-1β signaling were identified as upstream pathways (Supplemental Fig. 5), with MMP9 and CXCL-2 showing strong upregulation among inflammation-associated genes. Immunofluorescence analysis confirmed significantly more MMP9-positive cells in DBD than in DCD lungs (Fig. 2).
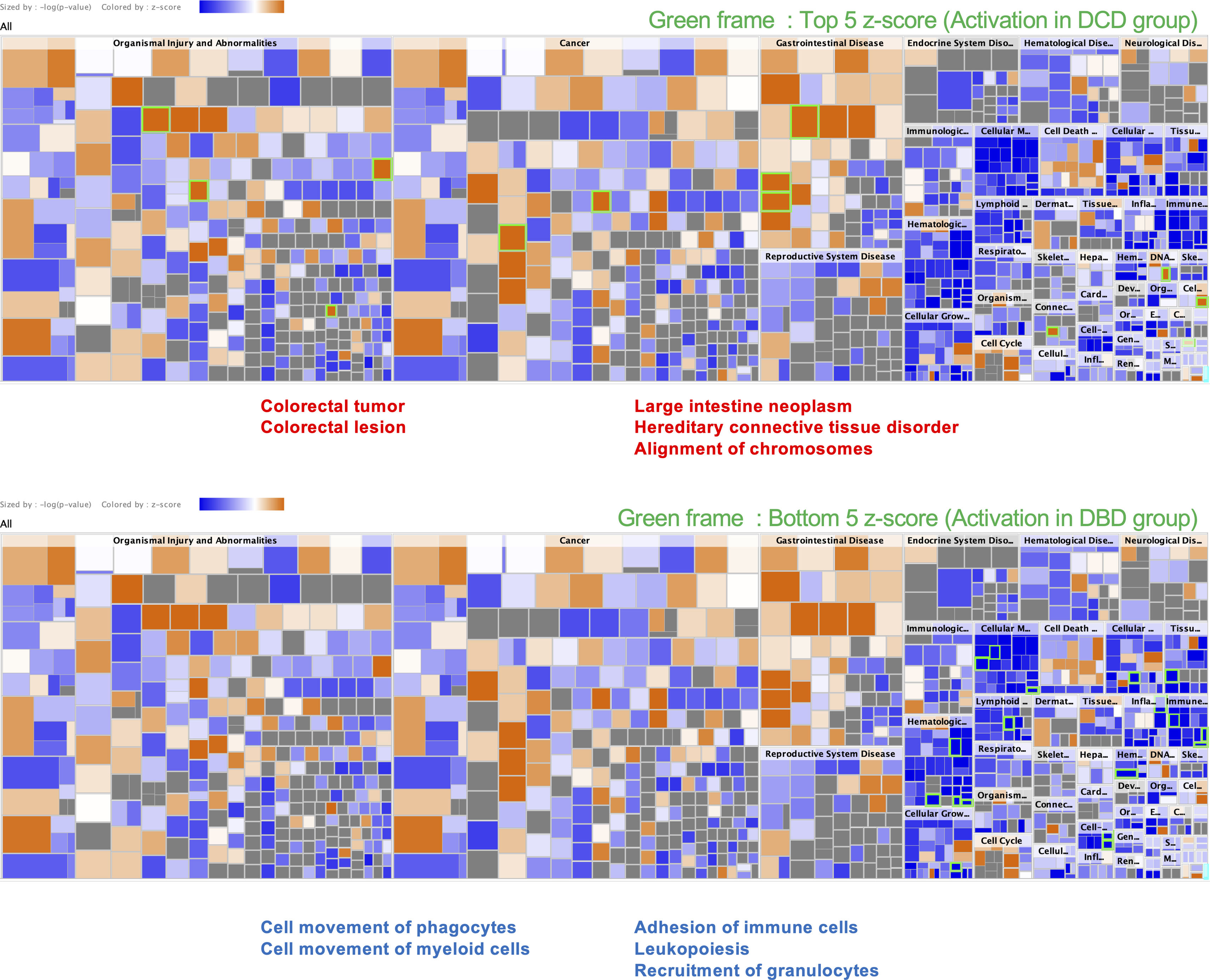
Disease and function analysis via IPA was conducted to investigate associations with previously published evidence (Supplemental Fig. 5). DCD lung had high homology to colorectal tumor and neoplasm, colorectal lesion, hereditary connective tissue disorder, and alignment of chromosomes. On the other hand, DBD lungs had high homology to the cell movement of phagocytes and myeloid cells, adhesion of immune cells, leukopoiesis, and recruitment of granulocytes. Cancer research has recently advanced therapeutic approaches targeting DNA damage responses,18,19 which may also be applied to DCD lung transplantation.
To our knowledge, this is the first study to analyze clinical DCD lung transplant specimens using RNA sequencing and immunofluorescence validation. Kang et al. previously employed microarrays to reveal differences in gene expression profiles between DBD and DCD lungs pre-transplant, with a notable reduction in these differences post-transplant.4 Further, their recent study on pre-transplant DBD and DCD lungs identified distinct transcriptomic signatures: DBD lungs displayed elevated inflammatory cytokine expression, while DCD lungs had signatures linked to cell death, apoptosis, and necrosis.20 While consistent with our findings, our study provides the first RNA-sequencing and immunofluorescence validation data for DCD lungs.
In conclusion, clinical lung tissues from DCD lung transplants exhibit significantly different gene expression profiles compared to DBD transplants. A combination of histological and transcriptomic analyses highlighted upregulation of DNA damage and replication signaling in DCD lungs, identifying DNA damage and replication as potential therapeutic targets for WIRI in DCD lung transplantation.
Author's ContributionsConcept and design: MO and TS; Collection of tissue samples: ST and YK; Analysis and interpretation of data: MO, TS and ST; Drafting/writing of the manuscript: MO, TS, ST; Manuscript reviewing: ST and DG; Technical support: ER, AS, MJC, LH and AR.
Ethics StatementThis study was approved by the Ethics Committee of Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University Hospital, Okayama, Japan (approval number: 2105-039), Ehime University Hospital, Toon, Japan (approval number: 2406011), and Hospital Universitario Puerta de Hierro-Majadahonda (approval number: AD0021-A-2015).
Funding StatementThis research was funded by Grants-in-Aid for Scientific Research from Japanese Society for the Promotion of Science (20KK0203), and a research grant from the Strategic Action on Health 2021–2023 from Instituto de Salud Carlos III (dossier PI22-01592).
Conflict of InterestsThe authors declare no competing interests.