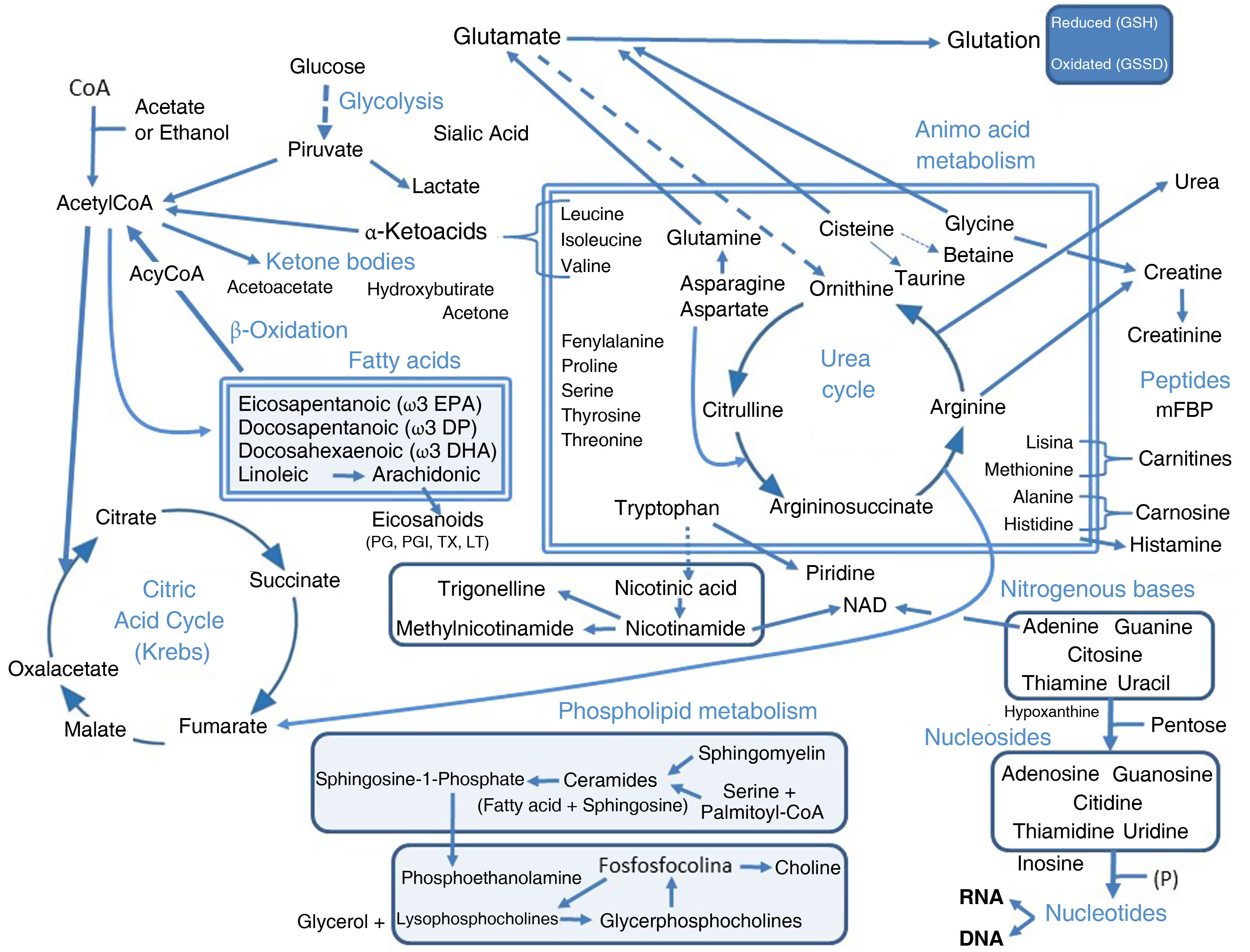
The clinical presentation of chronic obstructive pulmonary disease (COPD) is highly heterogeneous. Attempts have been made to define subpopulations of patients who share clinical characteristics (phenotypes and treatable traits) and/or biological characteristics (endotypes), in order to offer more personalized care. Assigning a patient to any of these groups requires the identification of both clinical and biological markers. Ideally, biological markers should be easily obtained from blood or urine, but these may lack specificity. Biomarkers can be identified initially using conventional or more sophisticated techniques. However, the more sophisticated techniques should be simplified in the future if they are to have clinical utility. The -omics approach offers a methodology that can assist in the investigation and identification of useful markers in both targeted and blind searches. Specifically, metabolomics is the science that studies biological processes involving metabolites, which can be intermediate or final products. The metabolites associated with COPD and their specific phenotypic and endotypic features have been studied using various techniques. Several compounds of particular interest have emerged, namely, several types of lipids and derivatives (mainly phospholipids, but also ceramides, fatty acids and eicosanoids), amino acids, coagulation factors, and nucleic acid components, likely to be involved in their function, protein catabolism, energy production, oxidative stress, immune-inflammatory response and coagulation disorders. However, clear metabolomic profiles of the disease and its various manifestations that may already be applicable in clinical practice still need to be defined.
Chronic obstructive pulmonary disease (COPD) is a highly prevalent disorder that carries a heavy social and economic burden.1,2 Diagnosis is based on a pertinent clinical history and the presence of chronic airway obstruction. Although some of the signs and symptoms of COPD are shared among all patients, its clinical presentation is highly heterogeneous and manifestations can be both pulmonary and extrapulmonary. Pulmonary expressions can range from emphysema to chronic bronchitis, presenting in different proportions in each patient. Progression also varies widely – some patients remain stable throughout their lifetimes, while others deteriorate rapidly. An important feature of COPD is the presence of intercurrent periods of worsening symptoms, known as exacerbations, that can be severe or very frequent. Attempts have been made in recent years to group different patient populations according to their clinical characteristics. These groups are called phenotypes or, when the emphasis is slightly more on therapy, treatable traits. Phenotypes are probably associated with distinct pathophysiological elements and comprise both the factors involved in the etiology and progress of the disease and specific biological mechanisms (endotypes). These concepts are already used in current COPD clinical guidelines,1,2 although, to date, they have not been sufficiently refined to be able to offer truly personalized medicine with highly precise management options for each patient.3 This knowledge gap contrasts with the emergence of new therapies, such as biological response modulators (known colloquially as ‘biologics’) aimed at modifying the mechanisms of disease presentation and/or progress.4
Although common elements clearly exist (e.g., inflammation or oxidative stress), it is easy to infer that the various mechanisms present in each patient and their interrelationship over time must be specific to some extent. This would have a significant impact on clinical practice, and would require the differentiation of therapeutic targets. It is therefore essential to determine the pathophysiological mechanisms present in each patient or group of patients. The use of both clinical (e.g., the presence and number of exacerbations, imaging or functional profiles) and biological markers seems to be the most appropriate way of identifying these mechanisms and assigning the COPD patient to a more specific subgroup. One such biomarker that has already helped personalize treatment to a somewhat greater extent is the number of eosinophils in peripheral blood.1,2 However, new elements remain to be incorporated into even more restrictive definitions of phenotypes/treatable traits and/or endotypes. The search for new biomarkers can be either targeted based on a specific hypothesis or ‘blind’, using massive screening processes without a pre-defined hypothesis. In addition to more traditional methods, scientific applications known as ‘-omics’ or ‘high-throughput’ have been widely used in the last decade. These methods select either both likely and or entirely unexpected molecules and metabolic pathways that may be potentially useful as clinical biomarkers and target molecules for new treatments.5
MetabolomicsThe various -omics are named after the substrate under study, for example, genomics, proteomics, or metabolomics. Metabolomics may be defined as the study of metabolites and the ramifications of the processes that generate them. A metabolite is an intermediate or final product of a biological process that serves as a clue to the most recent metabolic events. The metabolome is the collection of metabolites that exist in an organism, tissue, or organic fluid. Metabolites are generally molecules that weigh about 1500 daltons and comprise amino acids and peptides, components of nucleic acids, glucides, lipids and inorganic substances. Given that diseases such as COPD involve metabolic changes in the organism, derived mainly from the persistent inflammatory and catabolic state, it is reasonable to assume that these processes are reflected in the corresponding metabolites. Therefore, a metabolomic study could be useful both in the diagnosis of the disease and in differentiating its different phenotypes, and would lead to more precise and personalized management.6
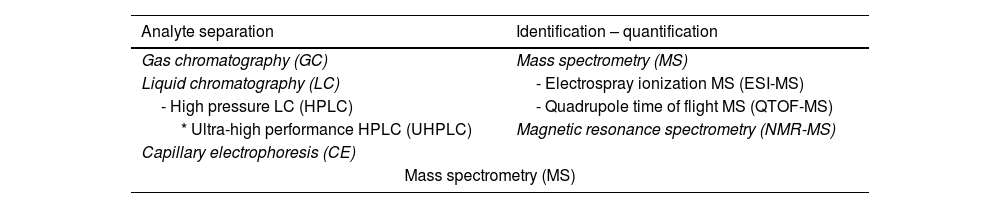
The elements that would be appropriate for a metabolomic study in COPD could be both local (such as lung tissue and airway secretions) and systemic (blood and urine). Systemic samples may be less organ-specific but they are much easier to obtain, so understandably most of the work has been done with these products. The techniques used in metabolomic analysis are primarily those that separate and subsequently identify and quantify ‘analytes’ (‘signals’) and their corresponding metabolites (Table 1). Gas chromatography (GC) is the technique most widely used to separate volatile compounds and other substances after derivatization.7 In contrast, hydrophilic or large ‘polar’ metabolites should be explored with other techniques such as high-pressure liquid chromatography (HPLC), which has a wider spectrum than GC.8 One version of HPLC is ultra-high performance liquid chromatography (UHPLC), which has many advantages, including smaller sample requirements, greater speed, and better resolution. Another technique is capillary electrophoresis (CE), characterized by its high metabolite discriminatory capacity.9 After the metabolites are separated using these techniques, mass spectrometry (MS) is used to identify and quantify them according to their analytes.7 MS can also be used as a single technique for separation, identification, and quantification. One type is electrospray ionization MS (ESI-MS), in which a very useful aerosol is generated for the detection of ions from macromolecules. HPLC, mentioned above, can also be linked to a quadrupole time of flight MS (QTOF MS). A more recent development is nuclear magnetic resonance (NMR) spectroscopy, which identifies metabolites without the need for prior separation, although relatively high proportions are needed.10 The results obtained with these techniques are usually expressed in relative terms, facilitating comparison with other samples in the same study but hindering direct comparisons with external results.
Techniques most commonly used in metabolomic studies.
| Analyte separation | Identification – quantification |
|---|---|
| Gas chromatography (GC) | Mass spectrometry (MS) |
| Liquid chromatography (LC) | - Electrospray ionization MS (ESI-MS) |
| - High pressure LC (HPLC) | - Quadrupole time of flight MS (QTOF-MS) |
| * Ultra-high performance HPLC (UHPLC) | Magnetic resonance spectrometry (NMR-MS) |
| Capillary electrophoresis (CE) | |
| Mass spectrometry (MS) | |
This review article discusses the most relevant findings in the literature on the metabolome of COPD patients. It is important to remember that the design of the studies, including the types of samples and the techniques employed, varies widely, so only an integrated overview can give an overall picture of the findings of this research.
Initial stages of exposureThe exposome includes all external and internal exposures of an individual from conception to death. Its relationship with genetic background may determine diseases, including COPD. In COPD, the influence of smoking and other pollutants is obvious, but other life habits and their time of onset, the environment, the development of both the immune system and the microbiota, and the various processes that occur in the body also play their part. The health impact of these factors changes dynamically over the life of an individual, and will be reflected in the metabolites generated at any given moment.11 This changing temporal relationship between COPD patients and their environment may also explain in part the different trajectories observed in this disease.12
Numerous studies have been published on the profound effects of smoking on metabolism. In an interesting study on blood drawn from smokers, Masthej et al. found that a network of 13 proteins and other metabolites (particularly amino acids and phospholipids) was inversely related to lung function.13 These results have been confirmed in other studies.14,15 Fibrinogen has also been associated with an increased risk of developing COPD among smokers.16
There are no specific metabolomic studies of the changes induced by wood or coal smoke, although reports are available on the effects of exposures to these particles. Nassan et al. found that exposure to particles smaller than 2.5μm (PM2.5), and in particular ultrafine particles, induces changes in phospholipid and glutathione metabolism that probably affect the inflammatory-immune response, oxidative stress, and the damage and repair of nucleic acids.17 Metabolomic studies of environmental pollution have been linked to oxidative stress, inflammation, and steroid metabolism in exposed individuals.11,17–21 It is also interesting to note that the metabolite profiles associated with environmental exposure appear to be different for each sex.22 In the case of COPD, these substances could possibly already be detectable both in the phases prior to the development of the disease and in its earliest stages.
Established lung disease: general aspectsNumerous metabolomic changes have been detected in COPD in both targeted and untargeted studies, but the results are still inconclusive. Most of these studies have been carried out on plasma or serum samples, not only because they are easy to obtain, but also because COPD is a disease with numerous systemic manifestations. Others have focused on samples from the respiratory system. In some of these publications, the degree of dilution is not known with certainty, compromising comparisons with other studies.23 Finally, some isolated studies in urine24–26 or even feces27 have been published.
The most relevant findings obtained in the various metabolomic studies carried out in COPD patients are discussed in this paper and the various phenotypes and other circumstances associated with the disease are detailed below. The results are summarized in Tables 2 and 3, and Fig. 1 has been provided for better understanding.
Most relevant associations of metabolites in COPD.
| AMINO ACIDS and derivatives16,24,26,28,33,36,41–43,48,56,58,60,61,63,64,66,67,70–72 |
| Alanine (b)71,72 (or ↑)73 |
| Arginine (b ↑)58,63,64 |
| Asparagine (b ↓)70 |
| Citrulline (b ↑)66 |
| Cysteine (BAL)28 (b ALT)71,72Glycine (b ↓)60,62 (BAL ↓)28 – dimethylglycine (b ↓)26,66 |
| Glutamine (b ↑)26,62,66,70 (EBC ↓)41 – Glutamate (b ↑-↓)58 |
| Histidine – methylhistidine (b ↑-↓)26,60,62,66,71,72 (or ↑)73 |
| Homocysteine (BAL ↓)28 |
| Isoleucine (b ↓ m)26,66 |
| Leucine (b ↓m)26,61,66 |
| Lysine (b NR)33.57 (EBC↓)43 |
| Methionine (b NR)71,72 |
| Ornithine (b ↑-↓)66 |
| Phenylalanine (b ↑)26,58,62.66 |
| Proline (b ↓ m) 26.61,66 (EBC ↓)43 |
| Proline (b ↓)60 (BAL↓)28 |
| Proline (b ↓)60 (EBC ↑)43 |
| Serine (EBC ↑)43 |
| Tryptophan (sp)36 |
| Tyrosine (b ↑)67 – Thyronine (EBC ↑)43 |
| Acetylpyridine [tryptophan] (E-nose ↓)48 |
| Carnitines and acylcarnitines [derived from lysine-methionine] (b ↑)24,56,58,67 |
| Carnosine (ur ↑)73 |
| Creatine [glycine and arginine] (b ↓)60 and creatinine (b ↑)16 (ur ↓)73 |
| Cysteinsulphonic acid [cysteine oxidation] (b ↑)16Glutathione (sp ↑)33 |
| Histamine [amine derived from histidine] (b ↑)60 |
| Hydroxyphenyl lactate [tyrosine derivative] (b ↑)67 |
| Methylisobutyric acid – methylisobutarate [leucine] (E-nose ↑)48 |
| PROTEINS, PEPTIDES and derivatives16,31,32,34,65,67 |
| Cyclooxygenase-2 (sp ↑)34 |
| CYTOCHROME P450 (BAL, NR)31 |
| Fibrinogen Peptide B [mFPB] (b ↑p)16 |
| Glycoprotein GlycA (b ↑)65 |
| HDL lipoproteins (b ↑)65 |
| Lipooxygenase -5 [5-LOX] (b ↓)31 |
| Methylurate (b ↑)67 |
| Myeloperoxidase [MPO] (sp ↑)32 |
| Superoxide dismutase [OD] (sp ↑)32 |
| NICOTINAMIDE and NICOTINIC ACID Metabolism28 |
| Nicotinamide (ur ↓)73 |
| p-Cresol (BAL ↓)28 |
| LIPIDS and derivatives24,26,28,29,31,32,34,37,41,42.43,44,53–59,62,65–67,70 |
| Fatty acids and relatedacids24,31,34,37,42–44,53,54,56and58,67 |
| Arachidonic acid [AA, ω-6] (sp ↑)34 |
| Eicosatetranoic acid [ETA, ω3] (b ↓)31 |
| Eicosapentanoic acid [EPA, ω3] (sp ↓)34 |
| Docosapentanoic acid [DPA, ω3] (sp ↑)34 |
| Docosahexaenoic acid [DHA, ω3] (b ↓)67 (sp ↓)34 |
| Hydroxy-heptadecatrienoic acid [HHTrE] (b ↓)31 |
| Lauric acid (b ↑)67 |
| Linoleic acid [LA, ω-6] (BAL ↑)31 (sp ↓)34 |
| Linoleic acid [ALA and GLA, ω-3] (sp↓)34 |
| Diglycerides and triglycerides24,53,56–58 |
| Diverse diglycerides, not specified (b ↑-↓)56–58 |
| Various triglycerides, not specified (b ↑)24,53,57,58 |
| Glycerophospholipids16,24,26,28,32,41,53,54,56–59 |
| Cardiolipin [a phosphatidylglycerol] (BAL ↓)28 |
| Glycerophosphocholine (b ↑)26 |
| Glycerophosphoinositol (b ↑)16 |
| Lysophosphocholine (b ↑)58 and lysophosphatidylcholine (b ↑)58 |
| Phosphatidylcholines (BAL NR)28 (sp)54,57 |
| Phosphatidylethanolamines (BAL NR)28 |
| Phosphatidylserines (BAL NR)28 |
| Phosphocholines (b ↑-↓)24,26,53,54,57–59 |
| Phosphorylcholine (EBC↓)41 |
| Sphingolipids28,29,53–56,59 |
| Sphingosine-1-phosphate (lt ↑)29 |
| Sphyingomyelin (b ↓) 53–55 (BAL↓)28 |
| Ceramides29,53–56 |
| Ceramides, not specified (b ↓)53–56 (lt ↑)29 |
| Othereicosanoids24,31,37,53,54 |
| Prostaglandin D2 [PGD2] (sp ↑) – derivative tetranor-PGDM (sp ↓)37 |
| Prostaglandin tetranor derivative E1 [PGE1] [PGEM] (sp↓)37 |
| Prostaglandin E2 [PGE2] (sp ↑) – derivative tetranor-PGEM] (sp↓)37 |
| Prostaglandin-like E2 [8-iso-PGE2] (sp ↑)37 |
| Prostaglandin-like F2α [8-iso-PGF2α] (sp ↑)32,37 |
| Leukotriene D4 [LTD4] (sp ↑)37 |
| Leukotriene E4 [LTE4] (sp↑)37 |
| Thromboxane D2 [11-dehydro-TBX2] (sp ↑) 37 |
| Modified steroids and derivatives67 |
| Androsterone sulfate (b ↑)67 |
| Cholestenoate (b ↑)67 |
| Ketone or relatedbodies26,43,46,48,62,65,66 |
| Acetate → Acetone (EBC↑)43 (ur ↑)26 |
| Acetoacetate – acetic acid (b ↑)65 |
| Cyclopentanone (E-nose ↑)48 |
| Hexanone (EBC↓)46 (E-nose ↑)48 |
| Ketoglutarate (ur ↑)26 |
| Ketone bodies, not specified (b ↑)26,62,66 (EBC↑)45,46 (ur ↑)26,73 |
| Phenylacetyglycine (ur ↑)26 |
| GLUCIDES and derivatives16 |
| Myo-inositol [Vit B8] (b ↑)16 |
| Sialic acid (sp ↑)33 |
| NITROGENOUS BASES and related33,67 |
| Adenine (sp ↑)33 |
| Hypoxanthine and xanthine (↑b)67 (sp ↑)33 |
| NUCLEOSIDES and related33.67 |
| Adenosine – Methylthioadenosine (sp ↑)33 |
| Pseudouridine (b ↑)67 |
| ACIDS and SALTS, Citric Acid Cycle (Krebs)16,41–43,68,69 |
| Acetolactic acid – Acetolactate → Acetoin (EBC NR)42 |
| Fumaric acid-fumarate (b ↑p)16 |
| Lactic acid – lactate (b ↑p)68,69 (EBC↑)42,43 |
| Maleic acid – Maleate (b ↑p)68 |
| Pyruvic acid – Pyruvate (b ↑)68.69 (EBC↓)41 (ur ↑)26 |
| Propionic acid – Propionate (EBC↑)42,43 |
| Succinic acid – Succinate (b ↑p)68 (EBC↓)41 |
| ALDEHYDES AND ALCOHOLS4,42,45,48,67 |
| Acetaldehyde [derived from ethanol] (E-nose ↑)48 |
| Ethanol (NR EBC)42 |
| Glycerol (b ↑)67 – Glycerate (b ↓)67 |
| Propanol(EBC↓)45,46 (E-nose ↑)48 |
| Various aldehydes, not specified (EBC↑)44 |
| HYDROCARBONS45–48 |
| Benzene (EBC↓)45 (EBC ↑)47 |
| Decanes (EBC ↑)45 |
| Hepatriene (EBC ↑)47 |
| Hexane (EBC ↓)45 and Cyclohexane → Hexanone (EBC↑)46 (E-nose ↑)48 |
| Indol (EBC ↓)47 |
| Limonin (EBC↓)45 |
| Octane (E-nose ↑)48 |
| Pentene (EBC↓)45 → Cyclopentanone (E-nose ↑)48 |
| Tetradecane (E-nose↓)48 |
| Toluene – Hydroxytoluene (EBC ↑)47 |
| MISCELLANEOUS28,33,36,41,45,47,48,67,70 |
| Choline (EBC ↓)41 |
| Dodecalactone (E-nose ↓)48 |
| Hydroxy-indolacetic acid [HIAA] (sp ↓)36 |
| Seratonin (b ↑)67 |
| Trimethylamine (b ↑)62 (EBC ↓)41 |
| Vinyl acetate [VAM, xenobiotic] (EBC ↑)47 |
| Vinylpirazine (E-nose ↓)48 |
The arrows indicate a higher (↑) or lower (↓) concentration in COPD patients compared to healthy subjects. In case of conflicting results, the larger arrow in the direction reported by more authors is shown. Bibliographic references are given for each particular molecule or group of molecules.
Abbreviations: b, blood (plasma or serum); BAL, bronchoalveolar lavage; EBC, exhaled breath condensate; E-nose, exhaled air detected by electronic nose; lt, lung tissue; NR, direction of changes not reported by the authors; p, predictor of poor prognosis; sp, sputum; ur, urine.
Most relevant associations of blood metabolites with the most accepted phenotypes described for the disease.
| (a) EMPHYSEMA13,55,58,60,62,66,78,79 |
| AMINO ACIDS, PEPTIDES and derivatives Arginine, Alanine, Asparagine, Citrulline, Phenylalanine, Glycine, Glutamine, Histidine, Lisine, Proline, Serine, Thyrosine, Triptophan and Valine |
| Aminoadipic acid and Aminoadipate (synthesis of lysine), Betaine, Carnitine, Creatine, Pyroglutamate and Sarcosine |
| LIPIDS and derivatives |
| - Fatty acids: Myristoleic acid |
| - Diglycerides: Palmitoyl-linoleoyl-glycerol |
| - Glycerophospholipids and derivatives: Stearoyl-linoleoyl-glycosylphosphatidylinositol, Stearoyl-arachidonyl-glycosylphosphatidylinositol, Palmitoyl-docosahexanoyl-glycosylphosphatidylethanolamine and Palmitoyl-linoleoyl-glycosylphosphatidylinositol |
| - Sphingomyelins |
| - Ketone bodies: Hydroxybutyrate |
| GLUCIDES: Glucose |
| NITROGENOUS BASES and derivatives: Adenine, aminoisobutyric acid (BAIBA, thiamine derivative) |
| NUCLEOSIDES: Ribosyl-Imidazolacetate |
| (b) LOW WEIGHT and CACHEXIA62 |
| AMINO ACIDS, PEPTIDES, PROTEINS and derivatives Glutamine, Glycine, Isoleucine, Methionine and Valine |
| Aminoisobutyrate (Thiamine and Valine catabolism) |
| HDL lipoproteins |
| LIPIDS and Ketone Bodies: Acetate, hydroxybutyrate |
| GLUCIDES: Glucose |
| ACIDS and SALTS, citric acid cycle and/or lipid β-oxidation: Pyruvate |
| MISCELLANEOUS: Ascorbic acid and nicotinic acid |
| (c) FREQUENT EXACERBATOR PHENOTYPE14,15,55,83 |
| AMINO ACIDS, PEPTIDES and derivatives: Alanine, Arginine, Leucine, Isoleucine, Proline, Serine, Threonine, Tryptophan and Valine |
| Betaine and aminoacyl-tRNA |
| (D) EOSINOPHILIC PHENOTYPE AND ASTHMA-COPD OVERLAP SYNDROME (ACOS)89,92 |
| AMINO ACIDS, PEPTIDES, PROTEINS and derivatives Glutamine-glutamate, Serine, Threonine and Valine |
| Betaine, IL-1β, IL-5, IL-6, IFN-γ, TGF-β and TNFα |
| LIPIDS |
| - Fatty acids: Stearic acid (octodecanoic) and linoleic acid |
| - Triglycerides: Palmitoylglycerol |
| - Eicosanoids: Various |
| - Modified steroids and derivatives: Cholesterol |
| GLUCIDES: D-mannose, glucose |
| ACIDS and SALTS, citric acid cycle: Citric acid-Citrate, Lactic acid-Lactate, and Succinic acid-Succinate |
| ALCOHOLS/AMINES: Ethanolamine |
Outline of the most relevant pathways, molecules, phenotypes, and other components associated with COPD detected in metabolomic studies (details in Tables 2 and 3).
Studies conducted on samples from the respiratory system itself best reflect events occurring in the lung and the airways.28
Lung tissue. Berdyshev et al., using liquid chromatography (LC)–MS, found that patients with mild-to-moderate COPD showed higher lung tissue levels of ceramides than control subjects, which was interpreted as an expression of tissue destruction.29 In contrast, patients with severe disease showed higher levels of a sphingolipid but lower levels of the above-mentioned ceramides.29
Bronchoalveolar lavage (BAL). Associations between lung function and some lipids (fatty acids, glycerophospholipids and sphingolipids), as well as various amino acids and some xenobiotics have been reported in the BAL of COPD patients by Halper-Stromberg et al.28 Other authors studying BAL have reported increases in various peptides that were also associated with functional impairment.30 Balgoma et al. found that lipids associated with linoleic acid and some derivatives of cytochrome P450 (involved in the synthesis of hormones, cholesterol, and vitamins) were closely related to lung function and the abundance of caliciform cells. In addition, a panel of 9 lipids differentiated between women smokers with or without COPD, although the comparisons were not useful in men.31
Induced sputum. Zhu et al. found reduced levels of several glycerophospholipid-related metabolites in the sputum of critically ill patients that probably impacted their level of oxidative stress, since these findings were also inversely related to levels of superoxide dismutase (SOD) and myeloperoxidase (MPO). SOD and MPO emerged from a total of more than 500 metabolites as being differentially expressed in moderate and severe COPD.32 Esther et al. found increases in sialic acid, glutathione (essential in redox balance), nitrogenous bases and nucleosides in the sputum of COPD patients.33 Also of interest is the study by Van der Does et al. who observed reduced levels of fatty acids and their derivatives in the sputum of patients with stable COPD, along with increases in arachidonic acid and mediators of cyclooxygenase-2.34 In the case of arachidonic acid, higher levels also appear to depend on sphingolipid-related pathways,35 with consequences for eicosanoid synthesis. In another interesting study, Yan et al. observed changes in tryptophan metabolism in sputum from COPD patients, due to a reduction in hydroxy-indolacetic acid (serotonin metabolism).36 Other molecules, such as some eicosanoids (prostaglandins, leukotrienes, and thromboxanes), have shown significantly higher levels in the sputum of COPD patients compared to controls.37
Exhaled breath condensate. Exhaled breath condensate has great potential value in the study of the ‘deep lung’. However, this sample is still rather difficult to obtain and analyze. Advances are needed to resolve problems in determining the real concentrations of each compound, whether volatile (gaseous at room temperature) or non-volatile (both organic and inorganic molecules).38–40 In one of the first published studies, de Laurentiis et al. found that patients, in contrast to healthy individuals, showed reduced levels or absence of some molecules involved in the citric acid cycle, amino acids, such as glutamine (synthesis of proteins and glutathione, and acid buffer), glycerophospholipid components, and choline.41 The authors reported years later that patients and controls also differed with respect to other markers associated with the citric acid cycle, fatty acids, and amino acids.42 These results were largely confirmed and even expanded by Bertini et al.43 and Bregy et al.44 Cazzola et al. analyzed various volatile compounds and reported differences between patients with COPD and controls for some hydocarbons, including many aromatic types, and some isolated xenobiotic compounds.45 These results are very similar to those reported by Westhoff et al.46 and Gaida et al.47
Electronic nose (eNOSE) exhaled breath analysis. This technology identifies compounds mainly linked to odors. Rodriguez-Aguilar et al., for example, reported that, compared to healthy controls, COPD patients have different levels of certain alcohols and their acetaldehyde derivatives, various hydrocarbons, amino acid derivatives, and ketone bodies.48
Systemic samplesBlood. Samples for analysis include serum or plasma, differentiated mainly by the presence of coagulation factors in the latter plasma. Numerous studies have been carried out using these samples, including some from very large multicenter projects such as ECLIPSE, COPDGene, and SPIROMICS,49–52 and other smaller investigations.
Chen et al. conducted a very interesting study comparing non-COPD smokers with COPD smokers, showing differences in 23 metabolites among the latter. This helped generate a highly predictive model that included glucides, glycerophospholipids, various peptides, including fibrinogen peptide B or modified fibrinogen peptide B, and amino acid oxidation products.16
More specifically, significant changes have been observed in several blood lipids. Some authors have described reduced serum or plasma sphingolipids in COPD patients, and lower levels of the previously mentioned ceramides among persistent smokers.53–55 Similarly, Gillenwater et al. revealed a strong association of disease with the network in which these 2 metabolites were found,56 and also reported that both diglyceride and some glycerophospholipid levels are reduced in patients.56 Reduced levels of phosphatidylcholines in patients were also confirmed in other studies.54,57 In contrast, other glycerophospholipids and associated molecules (such as phosphocholine, a precursor to phosphatidylcholine) and triglycerides appear to be increased.24,26,53,54,57,58 Liu et al., using ESI-MS, also demonstrated changes in various phospholipids, which in some cases were higher and in other cases were lower in patients compared to controls.59
Polyunsaturated fatty acids (PUFA) are also reduced in COPD, although monounsaturated fatty acids appear to be elevated, and changes in eicosanoids are also observed.24,53,54 Balgoma et al. observed that, compared to controls, COPD patients showed higher serum levels of CYP oxidase derivatives and some platelet acids derived from arachidonic acids, along with a decrease in lipooxygenase that transforms fatty acids into leukotrienes.31
The COPDGene and SPIROMICS cohorts reported that COPD patients showed changes in acylcarnitines (transport of fatty acids to the mitochondria for oxidation).56 Kim et al. and Callejon-Leblic et al. also reported similar findings in smaller patient cohorts.24,58
Protein metabolism changes have been reported in various amino acids and derivatives,24,26,56,58,60–66 with reduced levels mainly of branched-chain amino acids26,61 (more pronounced in severe patients),26,62,66 and others such as histidine and creatine (the latter derived from glycine and arginine). However, other amino acids (e.g., arginine and phenylalanine) or their derivatives (e.g., histamine, derived from histidine) show higher levels than in healthy controls,26,58,60,62,64,66 particularly during exacerbations.63 Some of these changes also correlate with levels of systemic inflammation markers.60,66 In parallel, several authors have shown higher levels of some HDL lipoproteins, GlycA glycoprotein (derived from acute phase proteins) and/or ketone bodies in COPD patients.62,65,66 Finally, changes have also been observed in modified fibrinogen peptide B that were directly related to the functional involvement of patients,16 possibly explaining some of their thromboembolic complications. If all the above is summarized in the protein metabolism pathways most closely associated with COPD, the specific amino acids cited and their oxidative processes seem to play a particular role, suggesting increased protein catabolism.33,57 Finally, in a large study conducted by Yu et al., up to 17 metabolites were found to be associated with COPD, including amino acids and their derivatives (e.g., ornithine, homocitrulline, and carnitines), various steroid lipids (e.g., androsterone and cholesterol-derived cholestenoate), serotonin, puric bases, and nucleosides.67 Xue et al. found elevated serum levels of maleate, pyruvate, and lactate in their patients, which also suggests oxidative metabolism problems in the Krebs cycle, with predominantly anaerobic glycolysis impacting on energy production,68 a concept also suggested by the results of Naz et al.69
Recently, Godbole et al. performed a study based on findings common to various studies conducted in the SPIROMICS and COPDGene cohorts in an attempt to generate a predictive model for functional impairment in COPD. Using a total of 132 metabolites, particularly amino acid and lipid-related ones, they were able to explain up to 65% of the variability of FEV1. However, this promising initial percentage was halved in the validation cohort.70 Huang et al. and Carpenter et al. also analyzed metabolomic results associated with environmental exposures, and confirmed that the disease is strongly related with alterations in the metabolism of amino acids and nucleosides.71,72
Urine. In one of the studies examining urine samples taken from smokers, some of whom were COPD patients, McClay et al. used NMR to determine that mainly trigonelline (a plant alkaloid), but also hippurate and formate, were associated with FEV1.25 All 3 metabolites are related to diet, but the latter 2 may also come from the intestinal microbiota.73 Wang et al. reported elevated urinary levels of ketone bodies and pyruvate, as well as carnosine dipeptide (antioxidant) and some amino acids, along with reduced levels of methylnicotinamide and creatinine.26
Animal modelsMultiple experimental COPD models have also contributed to the identification of metabolites possibly involved in this disease.74 This is the case of some fatty acids or prostaglandin PGE2,75 which were subsequently confirmed in patients with the disease. In addition, up to 70 characteristic metabolites have been identified in animal models of exacerbation.76 Animal models have also shown that some metabolites derived from smoking persist in blood for up to 2 months after stopping exposure.77 However, a detailed examination of the findings in COPD models goes beyond the scope of this review.
Severity of functional involvementSome associations between functional involvement and the metabolic profile of patients have already been mentioned.13,15,16,25,28 It should be added that several studies carried out on blood samples have reported that various sugars, amino acids and derivatives (e.g., γ-glutamylamino acids and aminoacyl-tRNA), diglycerides and phospholipids, nucleosides and trigonelline (derived from Vit B3) are associated directly or inversely with different pulmonary function variables.25,60,67,69 Indeed, pulmonary function generally correlates well with the pathways related to these substances, and with pathways of nitrogen metabolism, ATP-binding cassette (ABC) transporter and absorption of certain minerals.14,15,67 More recently, advanced machine learning methods (in particular, an approach called kernel machine regression) have been used to reinforce associations of this type, mainly with those of the aforementioned ABC transporters, the metabolism of certain amino acids, and those of the ligand-neuroactive receptor interaction.72
Phenotypes and COPDClassic phenotypes: emphysema and bronchitisA classic phenotype is emphysema patient. Several authors have observed the direct association of this structural alteration with various blood metabolites (Table 3), including citrate, various amino acids and associated molecules (such as betaine, protector of osmotic cellular stress), diglycerides, glycerophospholipids, and various sphingomyelins, ketone bodies, and some ribonucleosides.13,28,55,60,62,78,79 In contrast, other amino acids, sphingomyelins, ceramides, some peptides (e.g. sarcosine) or nitrogenous base derivatives (β-aminoisobutyric acid), and metabolites linked to protein acetylation have shown an inverse association with emphysema.62,66,78 It should also be noted that some metabolites show conflicting results, depending on the study. In another study, this time with BAL samples, Halper-Stromberg et al. found a high number of metabolites associated with emphysema, including those linked to amino acids, fatty acids and various phospholipids.28
Other pathways reported to be closely associated with emphysema include the Krebs cycle and oxidative phosphorylation,14,15 and perhaps with overproduction of free radicals. Three modules formed respectively by amino acids-phospholipids-bile acids-acyl cholines, amino acids-Krebs cycle-xenobiotics, and various steroids were found to be involved in the networks obtained by Gillenwater et al. from the COPDGene cohort.56 Unfortunately, the subsequent validation attempt in the SPIROMICS cohort did not fully confirm this data. In their machine learning analysis, Carpenter et al. obtained strong associations between the degree of emphysema and the ABC transporter networks, the metabolism of various amino acids, and the neuroactive ligand-receptor interaction.72 Most recently, Godbole et al. constructed a predictive model of emphysema and its intensity using the results of the SPIROMICS and COPDGene and 129 of the metabolites. This initially accounted for up to 60% of the variability, but this percentage fell to 17% in the validation cohort.70
Weight loss and muscle mass are common in COPD, especially in the emphysema phenotype and in advanced cases. In a separate investigation of this issue, Ubhi et al. reported a direct association between nutritional status and ascorbic acid (Vit C), a nicotinic acid ester (Vit. B3, in turn a precursor of adenine-nicotinamide dinucleotide or NAD), and HDL lipoproteins, and a negative association with glutamine. Extreme nutritional deficit or cachexia, meanwhile, were associated with decreases in branched-chain amino acids, ascorbate, pyruvate and glucose, and with increases in other amino acids and ketone bodies.62
The metabolic associations of the other classic phenotype, chronic bronchitis, have also been investigated in another study. Esther et al. reported elevated sialic acid levels in the sputum of these patients.33 However, other authors have not obtained such clear results when studying this metabolite in plasma,14 probably because this is a phenotype with more local respiratory system involvement. Callejon-Leblic et al. found differences between phenotypes classified as more pure emphysema versus chronic bronchitis: glucose serum levels were higher in emphysema phenotypes than in chronic bronchitis phenotypes, while myristoleic acid (ω-5) levels were lower; and in patients with emphysema certain amino acids and their metabolites were higher and others were lower.58 Tan et al. published an interesting study of the differences between emphysema patients who showed computed tomography findings of associated structural changes in the bronchial wall (a ‘mixed’ emphysema-bronchitis phenotype) compared to those without (more pure emphysema), noting that the mixed phenotype the former have lower levels of the amino acids glutamine and alanine than the latter pure emphysema phenotype.80
Muscle dysfunctionMuscle dysfunction, associated or not with weight loss and/or muscle mass, is another common manifestation in many patients with COPD.81 A study using NMR to investigate the plasma metabolomic profile of patients with muscle dysfunction reported reductions in alanine, isoleucine, and valine.82
Frequent exacerbatorsFrequent exacerbators also present distinct metabolic elements, as do subjects with particularly severe acute episodes. Increased levels of sialic acid, amino acids and derivatives (including glutathione) and nitrogenous bases were found in the sputum of frequent exacerbators included in the SPIROMICS cohort.33 Moreover, some of these biomarkers helped predict future exacerbations.33 Bowler et al. found that the systemic dysregulation of phospholipid metabolism observed in the plasma of COPD patients was mainly associated with glycosphingolipids in frequent exacerbators.55
In the SPIROMICS cohort, NMR analysis of blood samples showed that subjects with any exacerbation in the previous year present reductions in a large number of metabolites compared to other patients, primarily branched-chain amino acids and tryptophan.83 Cruickshank et al., in the COPDGene cohort, reported that subjects with frequent or severe exacerbations showed changes in various amino acids and derivatives (including aminoacyl-tRNA), sugars, and carnitines. The most disrupted pathways in these frequent exacerbators again involved these substances and those related to the metabolism of purines, ABC carriers, and absorption of certain minerals.15 In this same cohort of patients, Gillenwater et al. found that the amino acid derivative trimethyl-alanylproline and the already cited betaine were strongly associated with the number of exacerbations.14
Poor short-term prognosisA relatively rapid unfavorable course can also be considered a patient-specific profile. In this respect, compared to COPD patients with longer survival, those with reduced survival (LC–MS, LC–MS–MS and GC–MS) have shown a more marked reduction in branched-chain amino acids; higher levels of fructose and molecules linked to the citric acid cycle, whether standard (e.g., succinate, fumarate, malate and lactate) or variant forms (metabolism of glyoxylate and dicarboxylate); and well-defined alterations in the pentose-phosphate and glycerolipid pathways. These findings all imply particularly important problems in energy production,84 as also suggested by the networks created by Gillenwater and Naz.56,69 Patients with poor survival have shown elevated levels of a vasodilator bradykinin metabolite84 and of polypeptides associated with the cleavage of coagulation factor XII and fibrinogen, two components related with spontaneous clot formation.85 As already mentioned, a significant number of metabolites are associated with lung function and may be useful for biological monitoring that would help define different outcomes.
Another interesting topic is that of metabolic profiles in the earlier stages of the disease versus other more advanced stages, which may or may not always coincide with patient age. Indeed, age itself can shape different expressions of the disease. In the above-mentioned publication by Xue et al., it was observed that changes in the Krebs cycle inferred from the study results were much more evident in older patients than in young people with COPD.68
Eosinophilic COPD and asthma-COPD overlap syndromeOne of the most clearly defined phenotypes or treatable traits in COPD is the eosinophilic phenotype, commonly characterized by eosinophil levels greater than 300 cells/μl. This group accounts for approximately 15–30% of patients.86,87 The various metabolomic studies conducted to date have often indistictly combined this phenotype with the asthma-COPD overlap syndrome (ACOS), so both are included in this section.
Some authors have specifically studied ACOS-related alterations in the metabolome, revealing interesting relationships between lung function in these patients and blood levels of glycids (glucose and mannose), amino acids (e.g. glutamine, serine or valine), lipids (cholesterol), and citric acid cycle components (citrate and succinate),88,89 urinary levels of histidine,90 and levels of certain amino acids (valine), fatty acids, ketone bodies, alcohols and lactic acid in exhaled air condensate.91 Furthermore, some serum metabolites, including those mentioned above, and various sugars, succinic acid and ethanolamine89 clearly differentiate patients with ACOS from the other COPD or asthma phenotypes.89 ACOS patients also appear to have greater dysregulation of various cytokines89 and eicosanoids.92 In line with this, it is generally accepted that patients with ACOS are characterized by a more activated metabolic state and greater energy consumption93,94 than patients with only asthma or COPD, which could partially explain a worse outcome.39,95
An interesting and related topic is the difference between the more pure forms of COPD and bronchial asthma phenotypes. Liang et al., using LC–MS, observed that the pure COPD phenotypes showed higher plasma levels of some amino acids (the 3 branched-chain amino acids and phenylalanine), fatty acids and derivatives (arachidonic acid, pyroglutamic acid, and indoxyl sulfate), certain xanthines and Krebs cycle components (succinate), and lower levels of other lipids (e.g. palmitic acid, a component of cell membranes and lung surfactant) and xanthines, nucleosides (such as inosine), and bilirubin.96 De Laurentiis et al., in a study of exhaled breath concentrate, found that COPD patients showed higher levels of some alcohols (ethanol and methanol) than asthma patients, and lower levels of acetone.97 These results are very similar to those more recently reported by Maniscalco et al.98 Urine samples could also be useful for the same purpose, as suggested by the work of Adamko et al. who used NMR to determine that, compared to COPD patients, asthma patients showed higher levels of glutamine, succinate (again), the nitrogenous base uracil (enzyme synthesis and carbohydrate metabolism), and pantothenic acid (formation of coenzyme A [CoA], involved in cellular respiration and synthesis of immediate principles). They also showed lower levels of arginine, dimethylamine (synthesis of NO), hydroxyisovalerate (metabolism of CoA), betaine, choline, and methylnicotinamide (synthesis of steroid hormones).99
The reverse process: clustering by endotypesSimilarly to searches for metabolic biomarkers for the different disease phenotypes, attempts have been made to directly generate endotypes that in turn reclassify the phenotypes. The recent report from Gillenwater et al. is interesting in this regard: 2 new endotypes were generated from various metabolites related to sphingomyelins.100
Influence of sexAs mentioned previously, both very large studies (SPIROMICS and COPDGene cohorts) and more modest ones have found lower serum carnitine levels in female patients than in both men with COPD and healthy women.56,69 These serum findings are consistent with BAL findings from Balgoma et al., in which a panel of lipid mediators (specifically, various fatty acids or thromboxane B2 [TX2]) seemed to differentiate between female smokers with or without COPD, although this was not the case in men.31 In addition, nitric oxide and arginine pathways have shown more changes in the serum of women with COPD than in men, which may be related to the presence of higher levels of nitrosative stress in women.69 In contrast, the predominantly altered pathways in men are those related to various lipids (fatty acids, ceramides, and sphingolipids).56 In the networks obtained by Gillenwater et al. from the metabolites mentioned in previous sections, the modules formed by amino acids-lysophospholipids-bile acids-acetylcholines and amino acids-Krebs cycle-xenobiotics also appear more relevant in men with COPD, whereas a steroid network is more relevant in female patients.56 These and other differences in both metabolic markers and the clinical presentation of respiratory diseases between male and females have been related both to hormonal issues and to exposure to noxious substances related to the cultural environment.101,102
COPD exacerbationsVery few studies have addressed the appearance and predictability of metabolic changes at the specific time of exacerbations. Mention has already been made of the article published by Esther et al., which made it possible to establish predictions about upcoming acute episodes.33 Regarding the exacerbations themselves, Zhou et al. reported slightly lower levels of the amino acid taurine and the dipeptide glutamyl-phenylalanine (indicative of probable catabolic protein activity) during these episodes compared with periods of stability, although no clear predictive values could be defined.57 Gulcev et al., using NMR, also observed reductions in tryptophan levels during the first few days of an exacerbation, which is likely to be the result of a parallel increase in a dioxygenase.103 Finally, Van der Does et al. reported increases in arachidonic acids and DPA, and cyclooxygenase-2 mediators in the sputum of patients experiencing exacerbations.34 In terms of exacerbation severity, the catabolic pathways appear to be the most affected in severe cases, implicating both sugars and amino acids (notably arginine and proline), fatty acids and sphingolipids, as well as nucleosides, all of which very likely impact energy generation, use and storage.15,39 Fortis et al. showed that patients requiring admission and non-invasive mechanical ventilation showed reduced levels of glycine and formate.104 Patients with COPD and pneumonia show decreased levels of citrate and some amino acids and derivatives (histidine, glutamine, proline, and phosphocreatinine).104 However, despite all these studies, there is no precise metabolic marker of either exacerbation or severe exacerbation.
COPD and the microbiotaThe main component of the human microbiota is the intestinal flora, which is composed of a wide variety of microorganisms. Evidence is gradually emerging to suggest that the microbiota plays a fundamental role in the development and maturation of the immune system, with potential effects on organs such as the lung.105 Bearing this and the probable interactions between the intestinal and pulmonary microbiota in mind, the ‘gut-lung axis’ has been mooted.106 An important issue to keep in mind is that an individual's microbiota is subject to variations over their lifetime, depending on internal and environmental changes (infections, drugs, diet).107–110
Fecal sample studies have identified several metabolites that are differentially expressed between COPD patients and healthy controls. This is the case with various catabolites from the three immediate principles.27 The metabolic effects of the flora that makes up the microbiome can also be studied indirectly in other biological samples. McClay et al. found some metabolites suggestive of intestinal origin (e.g., hypurate and formate) that were expressed in the urine of COPD patients but not in controls.25 The microbiota also seems able to influence the heterogeneous clinical characteristics of COPD. Some authors, such as Kotlyarov et al. have suggested that oscillations in the levels of short-chain fatty acids derived from the metabolism of intestinal microorganisms would influence the differential expression of the disease.111 Similarly, other authors have reported differences in the intestinal microbiota of patients with mild or moderate disease versus those with more severe disease. In more severe patients, fusobacteria and aerococci are more abundant, and specific findings are also observed for the eosinophilic phenotype, namely abundant intestinal bacteroides.112 Some differences are also observed in the respiratory microbiome, in which the most severe patients present an increase of pseudomonas,113 while eosinophilic patients show more diverse genera, with greater abundance of thermophilic streptococci and fewer gram-negative bacillus Ochrobactrum than non-eosinophilic phenotypes.113,114 The composition of the respiratory microbiota has also been associated with functional performance, bronchial hyperreactivity, and clinical expression, particularly with regard to the genera Streptococcus, Staphylococcus and Pseudomonas,115 which are also associated with changes in the levels of various phospholipids, polyamines and xanthines in BAL.116 Furthermore, an impoverished respiratory microbiome in which Haemophilus dominates has been associated with an increased risk of death in COPD.117 With regard to the relationship between exacerbations and the respiratory microbiome, viral and bacterial burdens clearly increase the probability of exacerbations. This phenomenon has been attributed not only to their obvious infective capacity but also to their potential to damage the bronchial epithelium, stimulating the secretion of mucins and releasing of proinflammatory cytokines and reactive oxygen species.118,119 In addition, frequent exacerbators show reduced bacterial diversity,120 with increased populations of Pseudomonas ssp.113
Questions to be clarifiedAs various reviews of the topic have recently pointed out, the greatest knowledge gaps in metabolomics applied to COPD currently pertain to factors such as age and sex, disease at different ages, severity over time, severity, progression, and exacerbations.39,121 Other significant issues are that the control populations are often very younger than patients, and that multiple publications have been generated from a few cohorts that, though large, include patients and controls with very specific profiles. Moreover, validation studies of the initially generated models have so far been less than encouraging, so the clinical applicability of metabolomics in COPD still requires further advances and refinements. However, its potential is enormous both for diagnosis and for better definition of disease endotypes and phenotypes.
In summary, multiple studies of the metabolome of COPD patients have been conducted using diverse techniques and both respiratory and systemic samples. Although there are still lacunae, the findings on the metabolism of amino acids, lipids (mainly phospholipids and eicosanoids), and nucleic acids are of exceptional value. These results reveal different aspects of the pathophysiology and heterogeneity of the disease, such as the effects of inflammation, oxidative stress, protein catabolism, energy production deficits, and potential coagulation disorders.
FundingFunded in part by: SEPAR 2019 (project grant) and 2021 (travel grant), SOCAP 2021 (travel grant) and ISCIII (FIS PI21/00785 and M-BAE BA22/00009).
Cesar J. Enriquez-Rodriguez has received predoctoral assistance from ISCIII (PFIS ref. FI22/00003) cofunded by the EU.
Conflict of interestsThe authors state that they have no conflict of interests.
We thank Prof. Eyal Gottlieb of the Rappaport Medical Research Institute of the Technion Institute, Haifa (Israel), for his help.