Bi-allelic pathogenic variants in the cystic fibrosis transmembrane conductance regulator (CFTR) gene are the cause of cystic fibrosis (CF).1 More than 2000 specific variants have been reported to date. Most of these alterations are detected in exons and exon-intron boundaries (splicing variants). Changes in the promoter region, full or partial gene deletions, and more recently, deep intronic splicing variants (DISV) account for the remaining cases. DISV are alterations in the DNA sequence of intronic regions that generate cryptic splicing sites that favour the transcription of intronic sequences (pseudoexons) in mRNA molecules. These pseudoexons serve as templates for the synthesis of dysfunctional proteins, such as the CFTR protein in the case of CF, or create premature stop codons in the pre-mRNA molecules that result in nonsense mediated decay. These pathogenic variants could account for a majority of the 2–5% of remaining unknown variants in both cystic fibrosis (CF) and CFTR-related disorders (CFTR-RD) patients. The study of intronic sequences requires complex and expensive technology.2 We implemented next-generation sequencing (NGS) to study CF patients in our Genetic Laboratory in 2014 and were able to identify DISV in the second allele of four historical clinical CF cases that have been followed-up in our CF Unit during the last decades.
In order to assess the role of DISV on CF diagnosis, we present 4 cases of patients with clinical suspicion of CF but with incomplete genotype, in whom we detected DISV by NGS technologies.3 We used the CFTR MASTR™ Dx kit (Multiplicom, Niel, Belgium) directed to the 27 CFTR coding exons, selected intronic regions or variants and part of the CFTR promoter region.4 Cases 1–3 were diagnosed as CF during childhood by clinical manifestations and positive sweat test. However, only one mutated allele was detected in each patient during infancy. Case 4 was followed–up in our Unit since the age of 19.
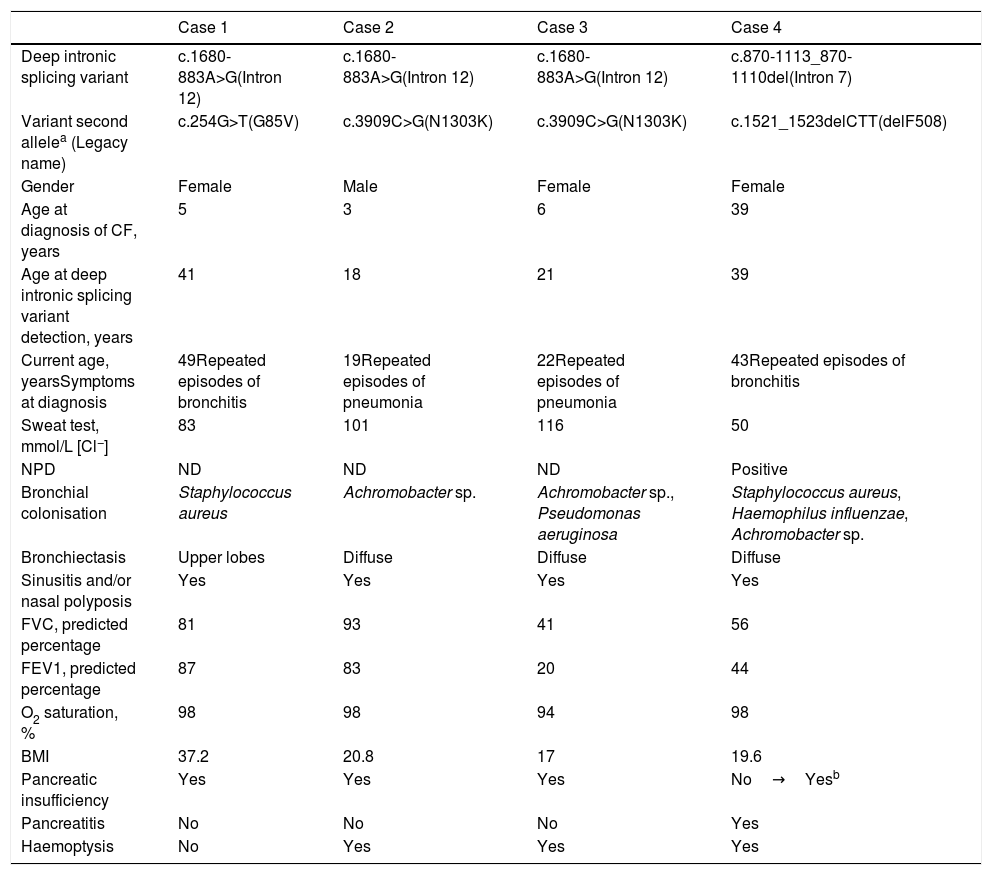
The clinical and demographic characteristics of the patients are shown in Table 1.
Clinical characteristics.
| Case 1 | Case 2 | Case 3 | Case 4 | |
|---|---|---|---|---|
| Deep intronic splicing variant | c.1680-883A>G(Intron 12) | c.1680-883A>G(Intron 12) | c.1680-883A>G(Intron 12) | c.870-1113_870-1110del(Intron 7) |
| Variant second allelea (Legacy name) | c.254G>T(G85V) | c.3909C>G(N1303K) | c.3909C>G(N1303K) | c.1521_1523delCTT(delF508) |
| Gender | Female | Male | Female | Female |
| Age at diagnosis of CF, years | 5 | 3 | 6 | 39 |
| Age at deep intronic splicing variant detection, years | 41 | 18 | 21 | 39 |
| Current age, yearsSymptoms at diagnosis | 49Repeated episodes of bronchitis | 19Repeated episodes of pneumonia | 22Repeated episodes of pneumonia | 43Repeated episodes of bronchitis |
| Sweat test, mmol/L [Cl−] | 83 | 101 | 116 | 50 |
| NPD | ND | ND | ND | Positive |
| Bronchial colonisation | Staphylococcus aureus | Achromobacter sp. | Achromobacter sp., Pseudomonas aeruginosa | Staphylococcus aureus, Haemophilus influenzae, Achromobacter sp. |
| Bronchiectasis | Upper lobes | Diffuse | Diffuse | Diffuse |
| Sinusitis and/or nasal polyposis | Yes | Yes | Yes | Yes |
| FVC, predicted percentage | 81 | 93 | 41 | 56 |
| FEV1, predicted percentage | 87 | 83 | 20 | 44 |
| O2 saturation, % | 98 | 98 | 94 | 98 |
| BMI | 37.2 | 20.8 | 17 | 19.6 |
| Pancreatic insufficiency | Yes | Yes | Yes | No→Yesb |
| Pancreatitis | No | No | No | Yes |
| Haemoptysis | No | Yes | Yes | Yes |
Abbreviations. CF: cystic fibrosis; NPD: nasal potential difference; ND: not done; FVC: forced vital capacity; FEV1: forced expiratory volume in the first second; BMI: body mass index.
The deep intronic variant c.1680-883A>G (rs1554388867, Legacy name c.1811+1637A>G, intron 12) was detected in cases 1–3, and the variant c.870-1113_870-1110del (rs397508809, Legacy name c.1002-1110_1113del, intron 7) in case 4. In the first three cases, CF was diagnosed during childhood owing to clinically compatible symptoms and positive sweat tests. The fourth case was initially classified as CFTR-RD at 19 years of age because of clinically compatible symptoms, an inconclusive sweat test (<60mmol/LCl−), and the detection of a single variant in the genetic analysis. CF was confirmed genetically 20 years later following the detection of a DISV in the second allele. This patient presented a severe phenotype with altered lung function, low body mass index (BMI), great extent of bronchiectasis, and a significant delay in diagnosis that could have negatively affected disease progression.
The deep intronic variant c.870-1113_870-1110del, which was found in case 4, is located in intron 7 of the CFTR gene and alters the mRNA splicing process creating a pseudoexon with a 101 nucleotide sequence between exons 6b and 7. This variant was initially identified in one patient with a clinically compatible phenotype and a negative sweat test, and in three Italian patients with a classic CF phenotype.5,6 Moreover, 17 patients with CF from other studies have also been diagnosed with this pathogenic variant.7–9 The patients have been reported as manifesting a wide spectrum of phenotypes including severe disease, delayed diagnoses, higher frequencies of diffuse bronchiectasis, with or without colonisation by Pseudomonas aeruginosa, pancreatic sufficiency (PS) that may progress to pancreatic insufficiency (PI) (as in our case 4 patient at 32 years old), and positive or inconclusive sweat tests. The variability of the phenotype has been related to levels of aberrant mRNA, which is affected by the expression of splicing factor SRp75. Decreasing levels of this factor have been shown to correct abnormal splicing of the variant.7
The deep intronic variant c.1680-883A>G is a more recently described variant located in intron 1210 that results in aberrantly spliced transcripts due to the inclusion of a pseudo-exon. Only three cases with this splicing intronic variant have been referenced in the literature and included in the CFTR-France Database.11 These three reported patients were diagnosed during childhood (2 months, 5 months, and 3 years) and were compound heterozygous with a positive sweat test, with or without PI, with respiratory symptoms at diagnosis, and without further follow-up data.11 Our three patients with this intronic variant (cases 1–3) now aged 49, 19 and 22 years respectively, are the only ones with available long-term follow up and progression data. They were diagnosed during childhood with positive sweat tests, are compound heterozygous for severe pathogenic variants and have PI, bilateral bronchiectasis, bronchial colonisation by Staphylococcus aureus, Achromobacter sp. or Pseudomonas aeruginosa, sinusitis and/or nasal polyposis, and preserved lung function, except for case 3, who had very impaired lung function and required a lung transplant at the age of 21. Our data support the involvement of this variant with a severe CF phenotype, in similar manner to c.1680-877G>T and c.1680-886A>G variants,12 and shows the long-term evolution of these patients when they are subjected to careful follow-up measures (case 3 was referred to our Cystic Fibrosis Unit just for lung transplant). Bonini et al.10 developed oligonucleotides that can correct aberrant splicing with the use of target site blocker treatment (TSB), opening the alternative of a tailored therapy for the causative defect. The use of other techniques capable of correcting aberrant splicing caused by other deep intronic variants have also been described recently.13–16
In conclusion, in CF cases with only one pathogenic variant detected, it is important to sequence the entire CFTR gene regions to confirm genetic diagnosis for management of the patient and to provide adequate genetic counselling. The increasing number of DISV reported10,17,18 will provide a better understanding of their pathogenic role in altering mRNA transcription using current technologies, such as NGS and new genome editing tools. This is essential in the present era for designing specific therapeutic approaches to correct the altered CFTR and move towards personalised medicine19 or 4P medicine (‘personalised’, ‘predictive’, ‘preventive’ and ‘participatory’).
FundingThis work has been partially supported by a grant of Fundación Mutua Madrileña. XIV Convocatoria de Ayudas a la Investigación en Salud. 2017.
Conflicts of interestThe authors have no conflict of interest to disclose.