While the molecular mechanisms of COPD pathogenesis remain obscure, there is mounting evidence supporting a key role for autoimmunity. Although human leukocyte antigens (HLA) alleles have been repeatedly associated with autoimmune processes, the relation between HLA and COPD remains largely unexplored, especially in Latin American (LA) populations. Consequently, this study aimed to investigate the presence of HLA class I and II alleles in COPD patients and healthy controls in a LA population with admixed ancestry.
MethodsCOPD patients (n=214) and age-matched controls (n=193) were genotyped using the Illumina Infinium Global Screening Array. The classic HLA alleles were imputed using HLA Genotype Imputation with Attribute Bagging (HIBAG) and the Hispanic reference panel. Finally, the distribution of HLA-DRB1 alleles was reexamined in 510 randomly recruited unrelated volunteers.
ResultsCODP patients showed a higher HLA-DRB1*01:02 allele frequency (6.54%) than healthy controls (3.27%, p=0.04, OR=2.07). HLA-DRB1*01:02 was also significantly associated with FEV1 (p=0.04) and oxygen saturation (p=0.02), and the FEV1/FVC ratio was higher in HLA-DRB1*15:01-positive patients (p=9×10−3).
ConclusionWe report an association among HLA-DRB1 alleles, COPD risk and pulmonary function parameters for the first time in Latin Americans. Since HLA-DRB1 genetic variability relates to the individual autoimmune response, these results support a role of autoimmunity in the pathogenesis of COPD.
Si bien los mecanismos moleculares de la patogénesis de la EPOC siguen sin ser claramente conocidos, cada vez existe más información que respalda que la autoinmunidad tiene un papel clave. Aunque los alelos de los antígenos leucocitarios humanos (HLA) se han asociado repetidamente con procesos autoinmunes, la relación entre los HLA y la EPOC permanece en gran parte inexplorada, especialmente en las poblaciones latinoamericanas (LA). En consecuencia, este estudio tuvo como objetivo investigar la presencia de los alelos de HLA de clase I y II en pacientes con EPOC y controles sanos en una población LA mestiza.
MétodosSe analizó el genotipo de pacientes con EPOC (n=214) y controles de la misma edad (n=193) utilizando el Illumina Infinium Global Screening Array. Los alelos clásicos de los HLA se imputaron usando la imputación del genotipo HLA con empaquetamiento de atributos (HIBAG, por sus siglas en inglés) y el panel de referencia hispano. Finalmente, la distribución de los alelos HLA-DRB1 se reexaminó en 510 voluntarios no emparentados reclutados al azar.
ResultadosLos pacientes con EPOC mostraron una mayor frecuencia de alelos HLA-DRB1*01:02 (6,54%) que los controles sanos (3,27%; p=0,04; OR=2,07). El HLA-DRB1*01:02 también se asoció significativamente con el FEV1 (p=0,04) y la saturación de oxígeno (p=0,02), y la relación FEV1/FVC fue mayor en los pacientes con HLA-DRB1*15:01 (p=9×10-3).
ConclusiónComunicamos una asociación entre los alelos HLA-DRB1, el riesgo de la EPOC y los parámetros de la función pulmonar por primera vez en latinoamericanos. Dado que la variabilidad genética de HLA-DRB1 se relaciona con la respuesta autoinmune individual, estos resultados respaldan el papel de la autoinmunidad en la patogénesis de la EPOC.
Chronic obstructive pulmonary disease (COPD) is the fourth leading cause of death worldwide and it is expected to become the third by 2020.1 In Latin American countries, the prevalence of COPD is 13.4% and its in-hospital mortality rate ranges from 6.7% to 29.5%.2 In Chile, respiratory diseases are the third cause of death and, amongst them, COPD accounts for 22%, being the second cause of decease.3
COPD is a complex disease where genetic variants, environmental factors and random events interact to trigger pathological pathways. Genetic susceptibility is evident in familial clustering.4 Specifically, population-based studies on families with respiratory disease have provided evidence for familial aggregation of spirometric parameters, and the heritability of lung function measures have been estimated to range from 10% to 80%.5,6 Large genome-wide analysis studies (GWAS) have increased our knowledge about the genetic risk factors for lung function impairment and COPD.7 However, most association studies trying to explain the genetic basis of COPD have been developed in Caucasian and Asian populations, showing some controversial results depending on the population analyzed.8 So far, only four GWAS assessing lung function or COPD-related phenotypes have been focused on Hispanic/Latino populations, reporting both some novel loci (in or near the genes KLHL7/NUPL2, DLG2, PDZD2 and PRDM15) and others previously identified in non-Hispanic samples.9–12 Moreover, single nucleotide polymorphisms (SNPs) identified through GWAS only explain a small percentage of the heritability of lung function parameters, such as forced expiratory volume in one second (FEV1, 9.6%), forced vital capacity (FVC, 6.4%) and FEV1/FVC ratio (14.3%).13
Although the molecular mechanisms of COPD pathogenesis are still unclear, mounting evidence supports that autoimmunity could play a role.14,15 Hence, the presence of autoimmunity-related responses elicited by T-helper type 1 (Th1) and Th17 cells have been reported in COPD patients,16,17 whereas abnormal levels of pulmonary and circulating autoantibodies have also been associated to COPD.18,19 Also, the results of some genetic studies have suggested a contribution of autoimmune responses to the development of COPD. For instance, Wain LV and colleagues have reported an association between decreased FEV1 and the HLA-DQB1/HLA-DQA2 region.20 In turn, a recent GWAS using samples from 18.335 Caucasian adults have identified a positive association of rs2074488, a SNP linked to the HLA-C gene, and COPD.21 Since human leukocyte antigens (HLA) alleles have been repeatedly associated with auto-immune diseases,22,23 these results reinforce the idea of an autoimmune component in COPD. Notwithstanding, the relation between HLA and COPD remains largely obscure, especially in Latin American (LA) populations.
To overcome this limitation, the present study aimed to analyze the presence of HLA class I and II alleles in COPD patients and healthy controls in a LA population with admixed ancestry.
MethodsStudy populationWe studied 214 COPD patients recruited at the Respiratory Service of the Hospital Regional de Talca, Chile, where they attended to undergo diagnostic tests after suspected COPD or for COPD monitoring visits. In parallel, 193 age-matched healthy controls were recruited at the same Hospital through a volunteer recruitment program. In order to rule out patients with asthma-COPD overlap syndrome, subjects with a history of asthma, rhinitis or any extra-pulmonary disease affecting lung function, and with positive bronchodilator test, FEV1 increasing by ≥12% and 200ml, were excluded from the study. In addition, 510 unrelated volunteers were randomly recruited among blood donors between January 2015 and May 2018 at Casa del Donante, Talca, Chile to reexamine the distribution of HLA-DRB1 alleles. 160 out of these 510 were submitted to HLA-A, -B and -C typing.24
Diagnostic evaluation of subjects was performed using GOLD criteria1 and medical history was considered standardizing clinical information. Pulmonary function – including measurements of FEV1, FVC and carbon monoxide diffusing capacity of the lung (DLCO) – was assessed in all subjects using standard procedures25 and equipment (Masterlab; Jaeger, Würzburg, Germany). Also, oxygen saturation was measured by pulse-oximetry (Ohmeda TuffSat, Soma Technology, Connecticut, USA). Body Mass Index (BMI) was calculated by dividing each person's weight in kilograms into their height in squared meters. Exercise capacity was determined with the distance walked in 6minutes test (6MWT). The amount of cigarette smoking history was measured by pack-years and cumulative exposure to biomass smoke was calculated and expressed as hour-years as previously described.26
Genotyping and imputationGenotyping was performed using the Illumina Infinium Global Screening Array (Illumina, California, USA).12 The classic HLA alleles at HLA-A, B, C, DPB1, DQA1, DQB1, and DRB1 were imputed using HLA Genotype Imputation with Attribute Bagging (HIBAG) with the Hispanic reference data set.27
Statistical analysesClinic and demographical data were expressed as mean±standard deviation. Comparisons between COPD patients and subjects were performed using the Student's t-test. SNPs that met the quality criteria of a minor allele frequency (MAF)>0.01, missingness<0.1, and/or Hardy–Weinberg equilibrium (HWE) p>0.001 were included in the association analyses. A total of 7257 SNPs was located on chromosome 6 (chr6: 28400538–33489882, hg19) (genotyping rate: 0.93). For single-variant association analysis, we used PLINK (v1.9) to perform logistic regression for binary phenotype (COPD and healthy controls).28 HLA association analysis was performed with the PyHLA software29 and R version 3.4.0. (https://cran.r-project.org/), using additive logistic regression models. Age, sex, and the first two principal components of a PCA based on genetic data, were used as covariates in all tests.
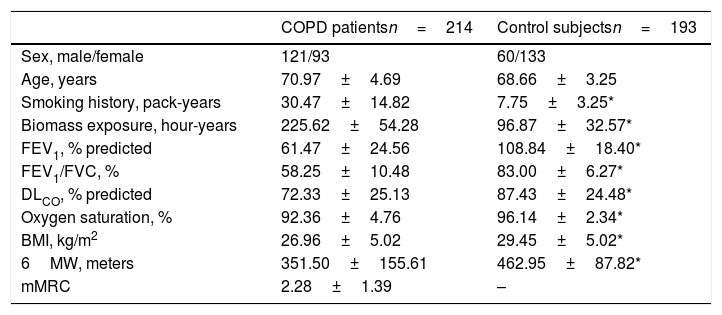
ResultsClinical and demographical findingsCOPD patients and controls were similar with regard to age, while smoking pack-years and biomass exposure were significantly higher in COPD patients (Table 1). As expected, lung function parameters were significantly reduced in COPD patients, as well as oxygen saturation and 6MWT. Control subjects exhibited a significantly higher BMI than COPD patients and, interestingly, both groups showed overweight (BMI=25.0 to <30).
Demographic and clinical data.
| COPD patientsn=214 | Control subjectsn=193 | |
|---|---|---|
| Sex, male/female | 121/93 | 60/133 |
| Age, years | 70.97±4.69 | 68.66±3.25 |
| Smoking history, pack-years | 30.47±14.82 | 7.75±3.25* |
| Biomass exposure, hour-years | 225.62±54.28 | 96.87±32.57* |
| FEV1, % predicted | 61.47±24.56 | 108.84±18.40* |
| FEV1/FVC, % | 58.25±10.48 | 83.00±6.27* |
| DLCO, % predicted | 72.33±25.13 | 87.43±24.48* |
| Oxygen saturation, % | 92.36±4.76 | 96.14±2.34* |
| BMI, kg/m2 | 26.96±5.02 | 29.45±5.02* |
| 6MW, meters | 351.50±155.61 | 462.95±87.82* |
| mMRC | 2.28±1.39 | – |
Data presented as mean±standard deviation, unless otherwise indicated. Definition of abbreviations: FEV1: forced expiratory volume in 1 second; FVC: forced vital capacity; DLCO: carbon monoxide diffusing capacity; BMI: body mass index; 6MW: 6minutes walking test; mMRC: modified Medical Research Council scale.
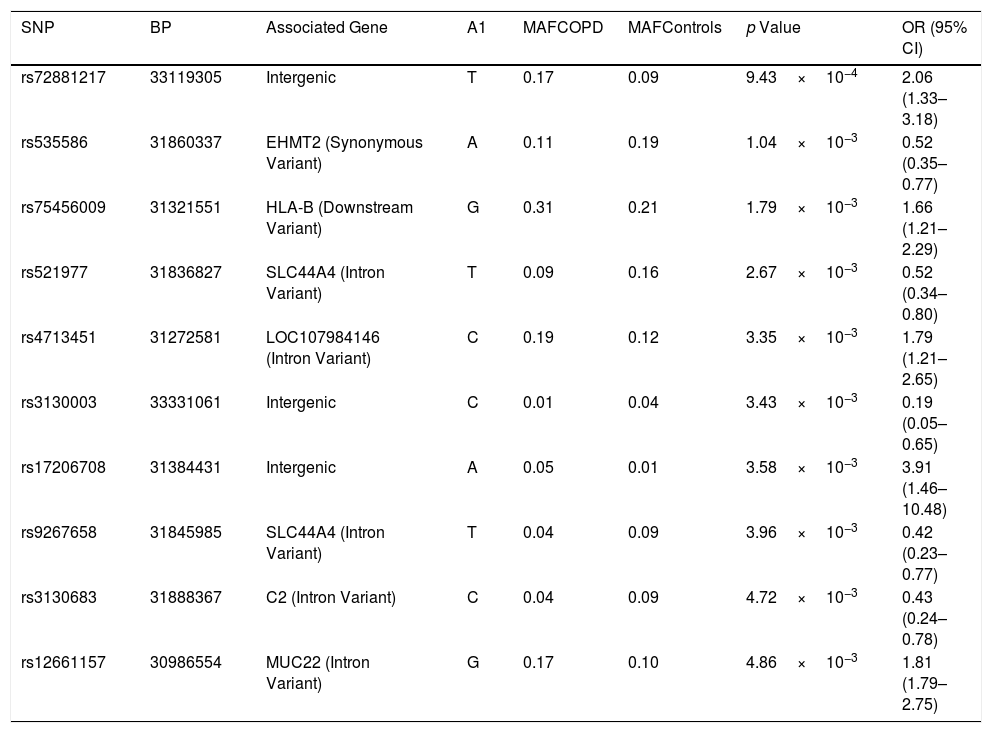
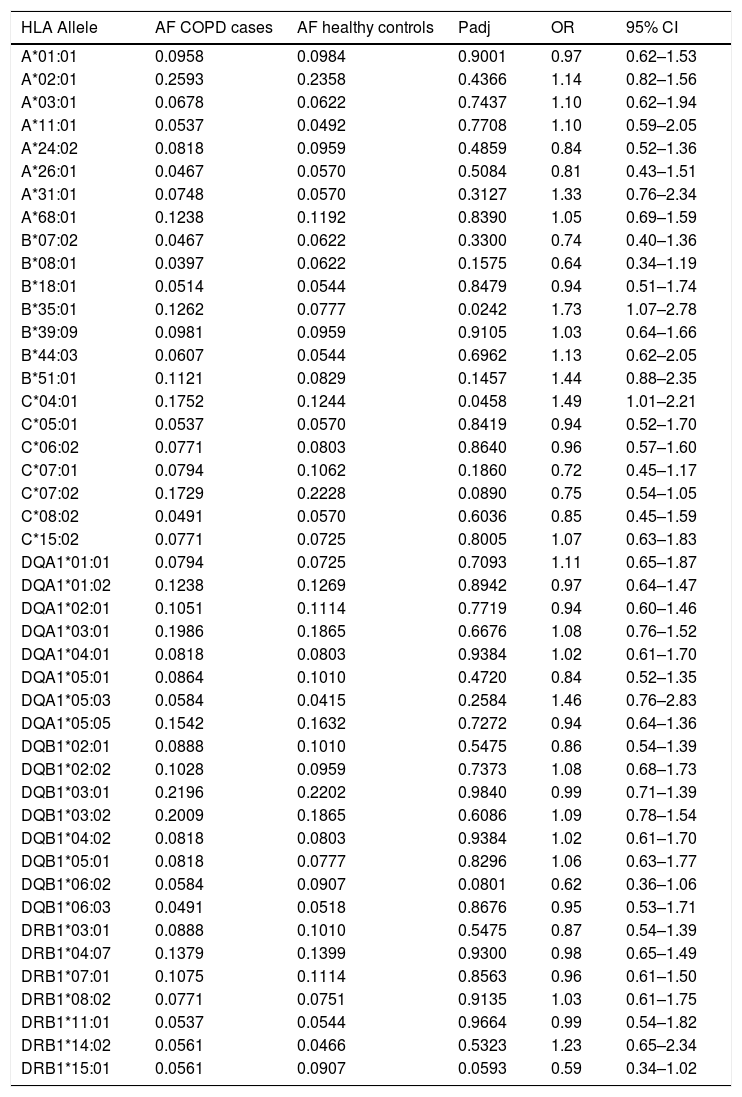
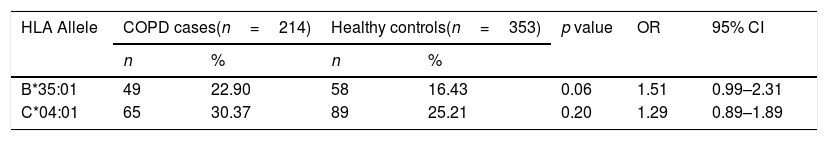
Of the 7257 SNPs genotyped, in the HLA region, ten markers showed suggestive associations (p<5×10−3) (Table 2, Supplementary Files 1). After HLA imputation, four HLA alleles showed a trend to association with COPD (Table 3; p<0.1). HLA-DRB1*15:01 was decreased in patients with COPD compared to healthy controls, although no significant difference was observed (OR=0.59, 95% CI 0.34–1.02). By contrast, the frequencies of HLA-B*35:01 and HLA-C*04:01 alleles were increased in COPD compared to controls (OR=1.73, 95% CI 1.07–2.78; and OR=1.49, 95% CI 1.01–2.21; respectively). The interaction test revealed that B*35:01 and C*04:01 were in LD in both COPD and controls (p=9.61×10−36 and p=5.67×10−23, respectively). Finally, we included in the analysis the HLA-A, -B, -C and -DRB1 typing performed in the Chilean population previously described by our group.24 No differences were observed in the distribution of HLA-B*35:01 and HLA-C*04:01 alleles among control subjects and patients with COPD (Table 4). On the contrary, the HLA-DRB1*01:02 allele frequency was increased in patients with COPD when compared with healthy controls (6.54% vs. 3.27%, p value<0.05, OR=2.07).
HLA SNP association values based on the allele frequencies characterized for the COPD patients vs. healthy controls.
| SNP | BP | Associated Gene | A1 | MAFCOPD | MAFControls | p Value | OR (95% CI) |
|---|---|---|---|---|---|---|---|
| rs72881217 | 33119305 | Intergenic | T | 0.17 | 0.09 | 9.43×10−4 | 2.06 (1.33–3.18) |
| rs535586 | 31860337 | EHMT2 (Synonymous Variant) | A | 0.11 | 0.19 | 1.04×10−3 | 0.52 (0.35–0.77) |
| rs75456009 | 31321551 | HLA-B (Downstream Variant) | G | 0.31 | 0.21 | 1.79×10−3 | 1.66 (1.21–2.29) |
| rs521977 | 31836827 | SLC44A4 (Intron Variant) | T | 0.09 | 0.16 | 2.67×10−3 | 0.52 (0.34–0.80) |
| rs4713451 | 31272581 | LOC107984146 (Intron Variant) | C | 0.19 | 0.12 | 3.35×10−3 | 1.79 (1.21–2.65) |
| rs3130003 | 33331061 | Intergenic | C | 0.01 | 0.04 | 3.43×10−3 | 0.19 (0.05–0.65) |
| rs17206708 | 31384431 | Intergenic | A | 0.05 | 0.01 | 3.58×10−3 | 3.91 (1.46–10.48) |
| rs9267658 | 31845985 | SLC44A4 (Intron Variant) | T | 0.04 | 0.09 | 3.96×10−3 | 0.42 (0.23–0.77) |
| rs3130683 | 31888367 | C2 (Intron Variant) | C | 0.04 | 0.09 | 4.72×10−3 | 0.43 (0.24–0.78) |
| rs12661157 | 30986554 | MUC22 (Intron Variant) | G | 0.17 | 0.10 | 4.86×10−3 | 1.81 (1.79–2.75) |
Only p values<5×10−3 are shown. A1: minor allele nucleotide; COPD: chronic obstructive pulmonary disease; MAF: minor allele frequency; SNP: single nucleotide polymorphism; OR: odds ratio; CI: confidence interval.
Frequency distribution of HLA alleles between COPD cases and controls.
| HLA Allele | AF COPD cases | AF healthy controls | Padj | OR | 95% CI |
|---|---|---|---|---|---|
| A*01:01 | 0.0958 | 0.0984 | 0.9001 | 0.97 | 0.62–1.53 |
| A*02:01 | 0.2593 | 0.2358 | 0.4366 | 1.14 | 0.82–1.56 |
| A*03:01 | 0.0678 | 0.0622 | 0.7437 | 1.10 | 0.62–1.94 |
| A*11:01 | 0.0537 | 0.0492 | 0.7708 | 1.10 | 0.59–2.05 |
| A*24:02 | 0.0818 | 0.0959 | 0.4859 | 0.84 | 0.52–1.36 |
| A*26:01 | 0.0467 | 0.0570 | 0.5084 | 0.81 | 0.43–1.51 |
| A*31:01 | 0.0748 | 0.0570 | 0.3127 | 1.33 | 0.76–2.34 |
| A*68:01 | 0.1238 | 0.1192 | 0.8390 | 1.05 | 0.69–1.59 |
| B*07:02 | 0.0467 | 0.0622 | 0.3300 | 0.74 | 0.40–1.36 |
| B*08:01 | 0.0397 | 0.0622 | 0.1575 | 0.64 | 0.34–1.19 |
| B*18:01 | 0.0514 | 0.0544 | 0.8479 | 0.94 | 0.51–1.74 |
| B*35:01 | 0.1262 | 0.0777 | 0.0242 | 1.73 | 1.07–2.78 |
| B*39:09 | 0.0981 | 0.0959 | 0.9105 | 1.03 | 0.64–1.66 |
| B*44:03 | 0.0607 | 0.0544 | 0.6962 | 1.13 | 0.62–2.05 |
| B*51:01 | 0.1121 | 0.0829 | 0.1457 | 1.44 | 0.88–2.35 |
| C*04:01 | 0.1752 | 0.1244 | 0.0458 | 1.49 | 1.01–2.21 |
| C*05:01 | 0.0537 | 0.0570 | 0.8419 | 0.94 | 0.52–1.70 |
| C*06:02 | 0.0771 | 0.0803 | 0.8640 | 0.96 | 0.57–1.60 |
| C*07:01 | 0.0794 | 0.1062 | 0.1860 | 0.72 | 0.45–1.17 |
| C*07:02 | 0.1729 | 0.2228 | 0.0890 | 0.75 | 0.54–1.05 |
| C*08:02 | 0.0491 | 0.0570 | 0.6036 | 0.85 | 0.45–1.59 |
| C*15:02 | 0.0771 | 0.0725 | 0.8005 | 1.07 | 0.63–1.83 |
| DQA1*01:01 | 0.0794 | 0.0725 | 0.7093 | 1.11 | 0.65–1.87 |
| DQA1*01:02 | 0.1238 | 0.1269 | 0.8942 | 0.97 | 0.64–1.47 |
| DQA1*02:01 | 0.1051 | 0.1114 | 0.7719 | 0.94 | 0.60–1.46 |
| DQA1*03:01 | 0.1986 | 0.1865 | 0.6676 | 1.08 | 0.76–1.52 |
| DQA1*04:01 | 0.0818 | 0.0803 | 0.9384 | 1.02 | 0.61–1.70 |
| DQA1*05:01 | 0.0864 | 0.1010 | 0.4720 | 0.84 | 0.52–1.35 |
| DQA1*05:03 | 0.0584 | 0.0415 | 0.2584 | 1.46 | 0.76–2.83 |
| DQA1*05:05 | 0.1542 | 0.1632 | 0.7272 | 0.94 | 0.64–1.36 |
| DQB1*02:01 | 0.0888 | 0.1010 | 0.5475 | 0.86 | 0.54–1.39 |
| DQB1*02:02 | 0.1028 | 0.0959 | 0.7373 | 1.08 | 0.68–1.73 |
| DQB1*03:01 | 0.2196 | 0.2202 | 0.9840 | 0.99 | 0.71–1.39 |
| DQB1*03:02 | 0.2009 | 0.1865 | 0.6086 | 1.09 | 0.78–1.54 |
| DQB1*04:02 | 0.0818 | 0.0803 | 0.9384 | 1.02 | 0.61–1.70 |
| DQB1*05:01 | 0.0818 | 0.0777 | 0.8296 | 1.06 | 0.63–1.77 |
| DQB1*06:02 | 0.0584 | 0.0907 | 0.0801 | 0.62 | 0.36–1.06 |
| DQB1*06:03 | 0.0491 | 0.0518 | 0.8676 | 0.95 | 0.53–1.71 |
| DRB1*03:01 | 0.0888 | 0.1010 | 0.5475 | 0.87 | 0.54–1.39 |
| DRB1*04:07 | 0.1379 | 0.1399 | 0.9300 | 0.98 | 0.65–1.49 |
| DRB1*07:01 | 0.1075 | 0.1114 | 0.8563 | 0.96 | 0.61–1.50 |
| DRB1*08:02 | 0.0771 | 0.0751 | 0.9135 | 1.03 | 0.61–1.75 |
| DRB1*11:01 | 0.0537 | 0.0544 | 0.9664 | 0.99 | 0.54–1.82 |
| DRB1*14:02 | 0.0561 | 0.0466 | 0.5323 | 1.23 | 0.65–2.34 |
| DRB1*15:01 | 0.0561 | 0.0907 | 0.0593 | 0.59 | 0.34–1.02 |
AF: allele frequency; Padj: p-values adjusted and corrected for FDR using the Benjamini–Hochberg test; OR: odds ratio; CI: confidence interval.
Frequency distribution of HLA alleles between COPD cases and controls.
| HLA Allele | COPD cases(n=214) | Healthy controls(n=353) | p value | OR | 95% CI | ||
|---|---|---|---|---|---|---|---|
| n | % | n | % | ||||
| B*35:01 | 49 | 22.90 | 58 | 16.43 | 0.06 | 1.51 | 0.99–2.31 |
| C*04:01 | 65 | 30.37 | 89 | 25.21 | 0.20 | 1.29 | 0.89–1.89 |
| HLA Allele | COPD cases(n=214) | Healthy controls(n=703) | p value | OR | 95% CI | ||
|---|---|---|---|---|---|---|---|
| n | % | n | % | ||||
| DRB1*01:02 | 14 | 6.54 | 23 | 3.27 | 0.04 | 2.07 | 1.04–4.10 |
| DRB1*14:02 | 23 | 10.75 | 48 | 6.83 | 0.08 | 1.64 | 0.97–2.77 |
| DRB1*15:01 | 24 | 11.21 | 88 | 12.52 | 0.64 | 0.88 | 0.55–1.43 |
OR: odds ratio; CI: confidence interval.
p values were determined by Fisher's 2-tailed exact test.
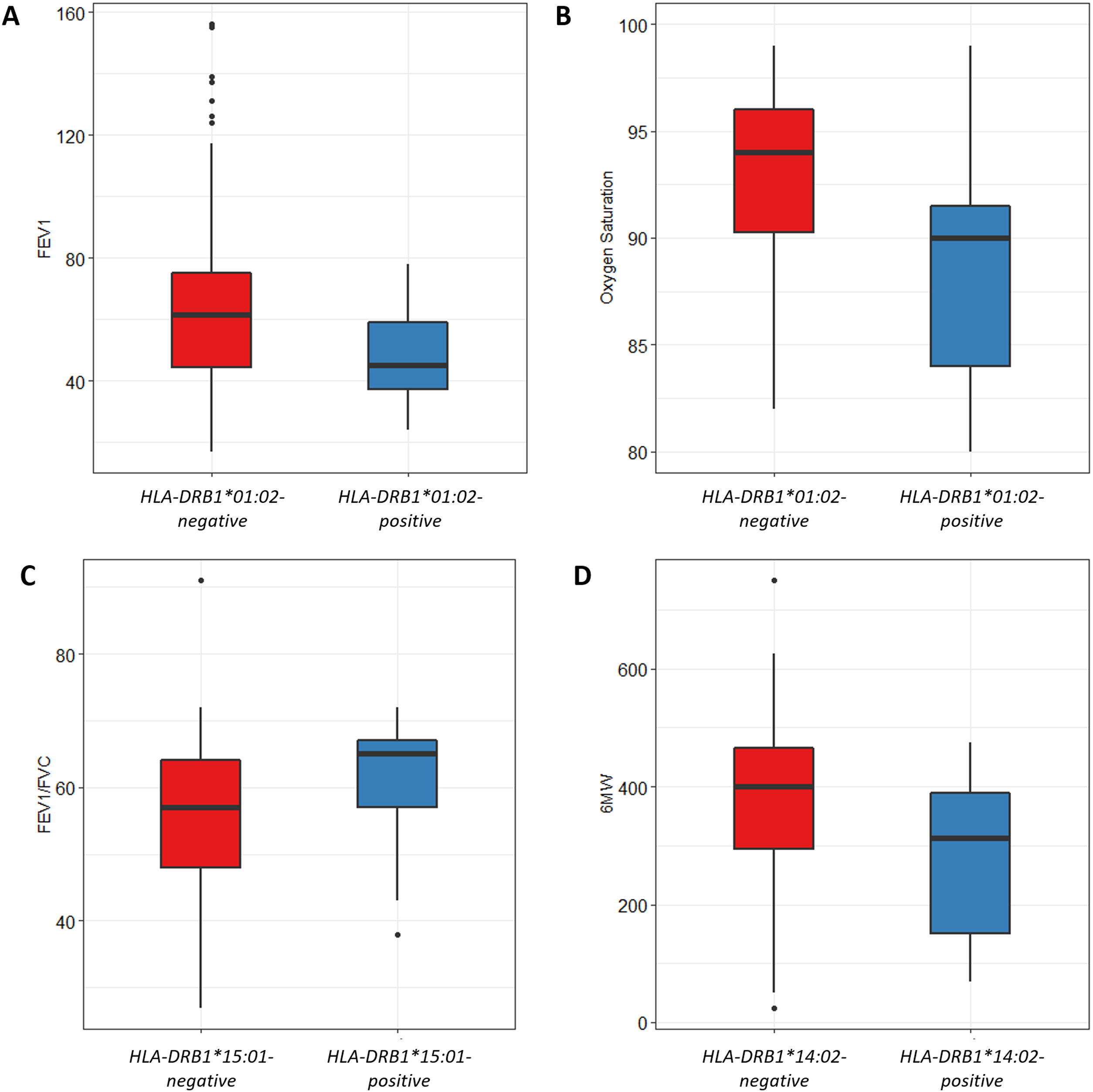
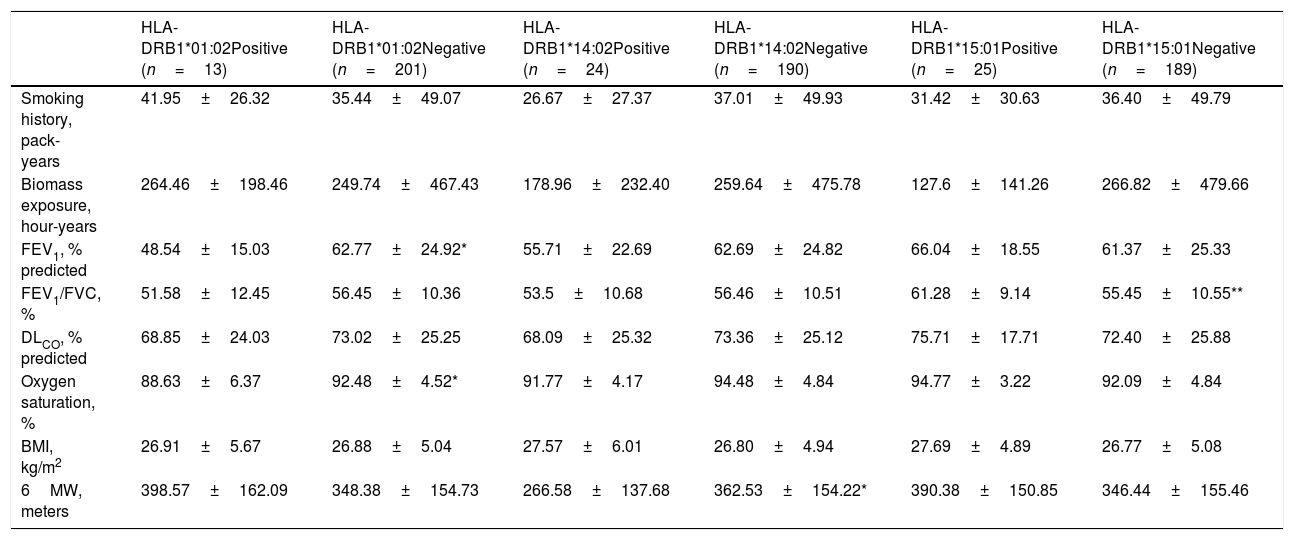
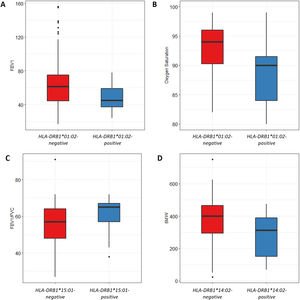
We also analyzed the possible influence of HLA class I and II alleles in pulmonary function, exercise capacity and oxygen saturation, in COPD patients (Table 5; Fig. 1). HLA-DRB1*01:02 was significantly associated with FEV1 and oxygen saturation (p=0.04 and p=0.02; respectively) (Fig. 1A and B). Moreover, FEV1/FVC ratio was higher among HLA-DRB1*15:01-positive patients compared with HLA-DRB1*15:01-negative patients (p=9×10−3) (Fig. 1C). Regarding exercise capacity, we observed that the median 6MWT value in the group of HLA-DRB1*14:02-negative patients was higher as compared with the HLA-DRB1*14:02-positive patient group (p=0.04) (Fig. 1D).
Genetic association between HLA-alleles, exposure to COPD risk factors and clinical parameters.
| HLA-DRB1*01:02Positive (n=13) | HLA-DRB1*01:02Negative (n=201) | HLA-DRB1*14:02Positive (n=24) | HLA-DRB1*14:02Negative (n=190) | HLA-DRB1*15:01Positive (n=25) | HLA-DRB1*15:01Negative (n=189) | |
|---|---|---|---|---|---|---|
| Smoking history, pack-years | 41.95±26.32 | 35.44±49.07 | 26.67±27.37 | 37.01±49.93 | 31.42±30.63 | 36.40±49.79 |
| Biomass exposure, hour-years | 264.46±198.46 | 249.74±467.43 | 178.96±232.40 | 259.64±475.78 | 127.6±141.26 | 266.82±479.66 |
| FEV1, % predicted | 48.54±15.03 | 62.77±24.92* | 55.71±22.69 | 62.69±24.82 | 66.04±18.55 | 61.37±25.33 |
| FEV1/FVC, % | 51.58±12.45 | 56.45±10.36 | 53.5±10.68 | 56.46±10.51 | 61.28±9.14 | 55.45±10.55** |
| DLCO, % predicted | 68.85±24.03 | 73.02±25.25 | 68.09±25.32 | 73.36±25.12 | 75.71±17.71 | 72.40±25.88 |
| Oxygen saturation, % | 88.63±6.37 | 92.48±4.52* | 91.77±4.17 | 94.48±4.84 | 94.77±3.22 | 92.09±4.84 |
| BMI, kg/m2 | 26.91±5.67 | 26.88±5.04 | 27.57±6.01 | 26.80±4.94 | 27.69±4.89 | 26.77±5.08 |
| 6MW, meters | 398.57±162.09 | 348.38±154.73 | 266.58±137.68 | 362.53±154.22* | 390.38±150.85 | 346.44±155.46 |
Genetic association between HLA-alleles and clinical parameters in COPD cases. (A and B) HLA-DRB1*01:02 was significantly associated with FEV1 and oxygen saturation (p=0.04 and p=0.02; respectively). (C) HLA-DRB1*15:01-positive patients exhibited a higher FEV1/FVC ratio than HLA-DRB1*15:01-negative patients (p=9×10−3). (D) HLA-DRB1*14:02-negative patients showed a higher 6MWT value (p=0.04).
The present study aimed to investigate the association of HLA class I and II alleles and COPD in the Chilean population, characterized by a large admixture ancestry. We show for the first time that HLA-DRB1*01:02 allele frequency is significantly increased in patients with COPD compared with healthy controls. Moreover, some HLA-DRB1 alleles correlated with clinical and pulmonary function parameters in COPD.
The HLA region plays a crucial role in numerous pathologies, as it accounts for 25% of known associations from the GWAS catalog (https://www.ebi.ac.uk/gwas/), especially with immune-related diseases. However, the relationship between HLA and COPD is unclear, since available data on this subject is scarce.30 In this regard, a previous study reported that the presence of alanine at amino acid position 57 (instead of aspartic acid, valine or serine) in HLA-DQβ1 was associated with decreased lung function.13 In turn, Faner and co-workers reported a higher prevalence of DRB1*14 in patients with severe airflow limitation and low DLCO.31 Likewise, here we report associations of HLA-DRB1 alleles with pulmonary function (HLA-DRB1*01:02 and HLA-DRB1*15:01), exercise capacity (HLA-DRB1*14:02) and oxygen saturation (HLA-DRB1*01:02), in COPD patients. Indeed, the HLA-DRB1 is the most polymorphic gene within the HLA-II region, and it has been frequently associated with autoimmune diseases such as rheumatoid arthritis (RA), spondylarthritis, systemic lupus erythematosus and multiple sclerosis, among others.32 Moreover, a meta-analysis performed in Latin American patients with different autoimmune-diseases revealed that specific HLA-DRB1 polymorphisms are shared between more than one of these conditions.33 Interestingly, it has been hypothesized that polymorphisms in HLA-II region could lead to the loss of immunological tolerance to self-antigens.34 In this regard, substantial evidence has shown the presence of self-antigens and autoantibodies in COPD patients.18,35,36 Hence, the HLA-DRB1 alleles showing association with COPD risk and lung function parameters could be involved in this auto-immune mechanism.
It is noteworthy that HLA-DRB1 shared epitope (SE) alleles (which encode a common amino acid sequence), are the most important genetic contributors for the risk of developing anti-citrullinated protein autoantibodies (ACPA).37 ACPA are different isotype autoantibodies that recognize the nonessential amino acid citrulline in proteins. Their presence has an important role in RA prognosis, since it is associated to severity of the disease.37 In this regard, it has been suggested that lungs may be initiating sites of the generation of ACPA.38 Although COPD is not a primary RA-related lung disease, a high prevalence of ACPA in COPD patients has been demonstrated.39 Moreover, it has also been reported that heavy smokers with COPD are more prone to ACPA production compared with heavy smokers without COPD.38 Hence, higher ACPA levels could increase the risk to develop COPD, even in the absence of RA. Indeed, citrullinated proteins are increased in patients with COPD and correlate with ongoing inflammation.40 Furthermore, it has been shown that anti-cyclic citrullinated peptide antibodies levels are higher in COPD patients exposed to biomass smoke.41 Although ACPA determination was not performed in our cohort, COPD patients exhibited a substantial exposure to biomass smoke. In this frame, it is conceivably that HLA-DRB1 alleles could contribute to ACPA generation as a result of immunization to newly synthesized citrullinated peptides.
On the other hand, we acknowledge some limitations, being the relatively small sample size and limited statistical power the most important. Hence, the associations found between COPD risk and HLA-B*35:01 and HLA-C*04:01 alleles disappeared when study population was increased. Moreover, the imputation of HLA alleles in LA populations may be controversial due to the scant information available and the complexity of the admixture. The fact that the number of males in the control group almost doubled that of the patients group could have also biased our results, since biological and behavioral/environmental gender differences have been recognized to influence COPD development.42 On the other hand, although we did not collect self-reported ethnicity, it has been shown that population from El Maule Region has a global ancestry estimate of 42.41% Native American, 55.62% European and 1.97% African.43 This is adds relevance to our analysis, since it is accepted that studies in admixed-ancestry populations could yield disease-associated loci that may have been missed due to allele frequencies disparities.12 Moreover, our findings are consistent with a plausible auto-immune process in COPD pathogenesis, which has been long recognized as a potential mechanism.
In conclusion, the present study shows an association among HLA-DRB1 alleles, COPD risk and COPD-related clinical parameters in an admixed LA population. Given that HLA-DRB1 gene is related to autoimmunity, our study supports the notion of an autoimmune process in the pathogenesis of COPD and justifies the need to develop more research on the association between HLA alleles and COPD risk.
Statement of ethicsThis research complies with the guidelines for human studies and was conducted ethically in accordance with the World Medical Association Declaration of Helsinki.
Authors’ contributionsStudy concept and design: JO, RDP; Data acquisition: RSS, SJ, JO; Data analysis: JO, RDP, HDH; Data interpretation: JO, RDP, MM; Funding acquisition: JO, RSS; Investigation: JO, RDP; Methodology: JO, RDP; Supervision: JO; Writing – original draft: JO, RDP; Writing – review & editing: JO, RDP, HDH, RSS, SJ, MM.
Funding sourcesFunding support for this study was provided by the Chilean National Science and Technology Fund (CONICYT), FONDECYT Project N° 11150022.
Conflict of interestMarc Miravitlles has received speaker fees from AstraZeneca, Boehringer Ingelheim, Chiesi, Cipla, Menarini, Rovi, Bial, Sandoz, Zambon, CSL Behring, Grifols and Novartis, consulting fees from AstraZeneca, Boehringer Ingelheim, Chiesi, GlaxoSmithKline, Bial, Gebro Pharma, CSL Behring, Laboratorios Esteve, Ferrer, Mereo Biopharma, Verona Pharma, TEVA, pH Pharma, Novartis and Grifols and research grants from GlaxoSmithKline and Grifols, all outside the submitted work.
The rest of the authors declare no conflicts of interest.
The authors would like to thank Mr. Felix Boekstegers, Mr. Justo Lorenzo Bermejo, Ms. Viviana Parra, Ms. Hanuxa Celedón and Mr. Adam Darski for their technical assistance. They also thank Marta Chmielewska for linguistic advice.