Molecular analysis of the gene that encodes alpha-1 antitrypsin (AAT; SERPINA1 gene) is the gold standard for the identification of allelic variants.1 The different molecular methods that can be used for this purpose include hybridization probes or HybProbes,2 which provide real-time PCR tracking. Once the amplification process is complete, these probes identify the genetic variants present in a particular region within the amplicon. This is a homogeneous genotyping test, that is to say, the entire process occurs in a single tube with no additional manipulation between the start of the test and the observation of the results. However, while it is a very reliable technique, errors may sometimes occur, especially in the interpretation of the results.3
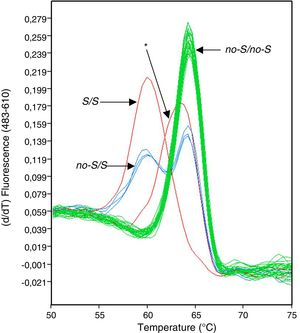
We performed an analysis of the prevalence of non-S/S and non-Z/Z variants of the SERPINA1 gene in a clinical population from La Palma (Canary Islands, Spain) by recruiting a series of 1510 patients regardless of the reason that led them to the pulmonology clinic. We identified 7 subjects in whom the peaks in the melting charts displayed by the HybProbe probes designed to identify the non-S/S variants showed a shift in respect of normal charts (Fig. 1). These 7 patients had been diagnosed with various respiratory diseases, such as diffuse interstitial lung disease, sleep apnea–hypopnea syndrome, and chronic obstructive pulmonary disease.
To rule out an error in the genotyping process due to differences in the saline concentration of the 7 DNA samples involved, these were prepared and analyzed again. In the new analysis, the real-time PCR genotyping platform software (LightCycler 480) continued to allocate these samples to a different genotype group than those defined by the standards, using the computer application's default threshold values for similarity and resolution. A review of the melting temperatures indicated a clear difference between the melting peaks of the 7 samples (average=63.21°C; SD=0.05) and the closest melting peaks from samples with a non-S/S genotype (mean=64.18°C; SD=0.26; n=71).
These results suggested that the SERPINA1 gene in those individuals contained a mutation other than the S variant (c.863A>T; p.Glu264Val) in the region covered by the genotyping probes. One candidate was the c.839A>T (p.Asp256Val) mutation, a severe deficiency variant with clinical implications that defines, depending on the genetic background, the alleles PI*Plowell, PI*Pduarte and PI*Ybarcelona.4–6 This possibility was ruled out when it was found that serum AAT levels of 7 patients were in the range of 93.5–167mg/dl. Despite these patients had not yet shown signs of severe AAT deficiency, we decided to continue investigating what appeared to be the most frequent rare genetic variant in our clinical sample. In this respect, the samples of 3 of the 7 patients underwent amplification of the coding region of the SERPINA1 gene and the corresponding introns, in the form of two overlapping amplicons, which were sequenced. This analysis revealed that the 3 patients were carriers of the c.840T>C (p.Asp256Asp) mutation in heterozygosis, specifically in the region to which the anchor probe used to detect the non-S/S variants was attached. Moreover, one of these patients was heterozygous for the c.774G>A (p.Lys236Lys) mutation. These mutant variants appear not to be deficiency variants, since the levels of AAT measured in PI*MMD566D patients (166 and 142mg/dl) and in the PI*SMD566D patient (125mg/dl) fit perfectly within the reference range for individuals with genotype PI*MM or PI*MS.7 This observation is predictable, since these are 2 mutations that do not change the protein sequence (synonymous mutations), and can also be expected not to affect the analysis of the phenotype of these patients by isoelectric focusing.
The c.840T>C mutation, in the normal M1-Val213 allelic background, in combination with the c.1093G>A (p.Asp341Asn) mutation make up the non-deficiency allele PI*Psaint albans.4 Since this second mutation was not found in the 3 patients analyzed, we ruled out the presence of the allele in these individuals. Since conventional PCR simultaneously amplified the 2 alleles of the SERPINA1 gene, we could not determine exactly in which normal allelic background the mutations detected in these patients were located. However, we could deduce which combination of these allelic backgrounds were present in each patient. Interestingly, the c.840T>C mutation in the M1-Ala213 background, or the combination of c.840T>C and c.774G>A mutations in the M1-Val213 background, have only been described to date in the sub-Saharan population from South Africa, where they reach allele frequencies of 10.4% and 4.3%, respectively.8 Although confirmation would require haplotype analysis, these sub-Saharan alleles might be found in the current population of the island of La Palma and, presumably, were incorporated via African slaves brought to the island after the mid-16th century.9
The authors would like to thank Grifols International S.A. for funding this research.
Please cite this article as: Hernández Pérez JM, Pérez Pérez JA. Las alteraciones en el pico de fusión de las sondas de hibridación usadas para el genotipado en la deficiencia de alfa-1 antitripsina no siempre implican errores. Arch Bronconeumol. 2019;55:338–339.