Bronchiectasis is characterized by chronic airway infection, persistent inflammation, and tissue damage, creating an environment portending toward antimicrobial resistance.1 While pathogens have historically been the focus, the growing recognition of the lung microbiome—including commensals and pathobionts—now add complexity to understanding resistance dynamics and therapeutic outcomes.2,3 The advent of next-generation sequencing technologies has transformed our ability to characterize resistance at the strain level and is increasingly being applied in epidemiological and clinical contexts.4 This has led to accelerated interests in pathogen genomic research, including microbiome analysis and its associated resistome. In some cases, in silico resistance prediction is supplementing traditional culture-based susceptibility testing, exemplified by its clinical application in Mycobacterium tuberculosis.5 However, unlike M. tuberculosis, whose closed pan-genome and mutation-driven evolution make resistance prediction more straightforward, pathogens with open pan-genomes such as Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli pose greater challenges due to extensive recombination and horizontal gene transfer, although bioinformatic prediction tools are now making encouraging progress.4,6 Critically, established resistance prediction tools and databases including ResFinder, the Comprehensive Antibiotic Resistance Database (CARD) and the Antibiotic Resistance Genes Database (ARDB) remain focused on established pathogens, limiting their ability to capture novel or commensal-associated resistance determinants, and therefore only offering a biased view of the broader resistome.7,8 Such challenges are further exacerbated in metagenomic studies of the lung microbiome, where diverse and poorly characterized species coexist alongside established pathogens, complicating resistance prediction at the community level.
Advances in metagenomics now enables functional characterization of the lung microbiome, offering insights into resistome structure.9 In bronchiectasis, antibiotic resistance emerges through stepwise mutations disrupting antibiotic–drug target interactions and the independent acquisition of exogenous resistance.4 Although initially identified in human pathogens, many horizontally acquired resistance determinants have origins in environmental or neighboring commensal species, which thus represent key microbial reservoirs of clinical resistance.8,10 Within the microbiome, the resistome therefore represents an integrated, adaptable functional substructure that responds to antibiotic pressures. Here, sensitive organisms decline, while resistant strains persist or even expand, reshaping the ecosystem based on functional composition. Resistome profiling may therefore offer a complementary view to taxonomic microbiome analysis providing distinct analytical insights.11 High-throughput sequencing—through short-read shotgun approaches and, increasingly long-read technologies—now permits resistome assessments at unprecedented scales.11,12 Advancing from inference, based on taxonomy or isolate-specific sensitivity, metagenomics offers a potential for therapeutic stratification, personalized antibiotic regimens and improved stewardship tailored to individualized resistance landscapes. Recognizing the resistome as a dynamic, functional feature of the lung, and not merely a reflection of pathogen burden, therefore represents a major shift in understanding of the bronchiectasis milieu and the treatment response1,8 (Fig. 1).
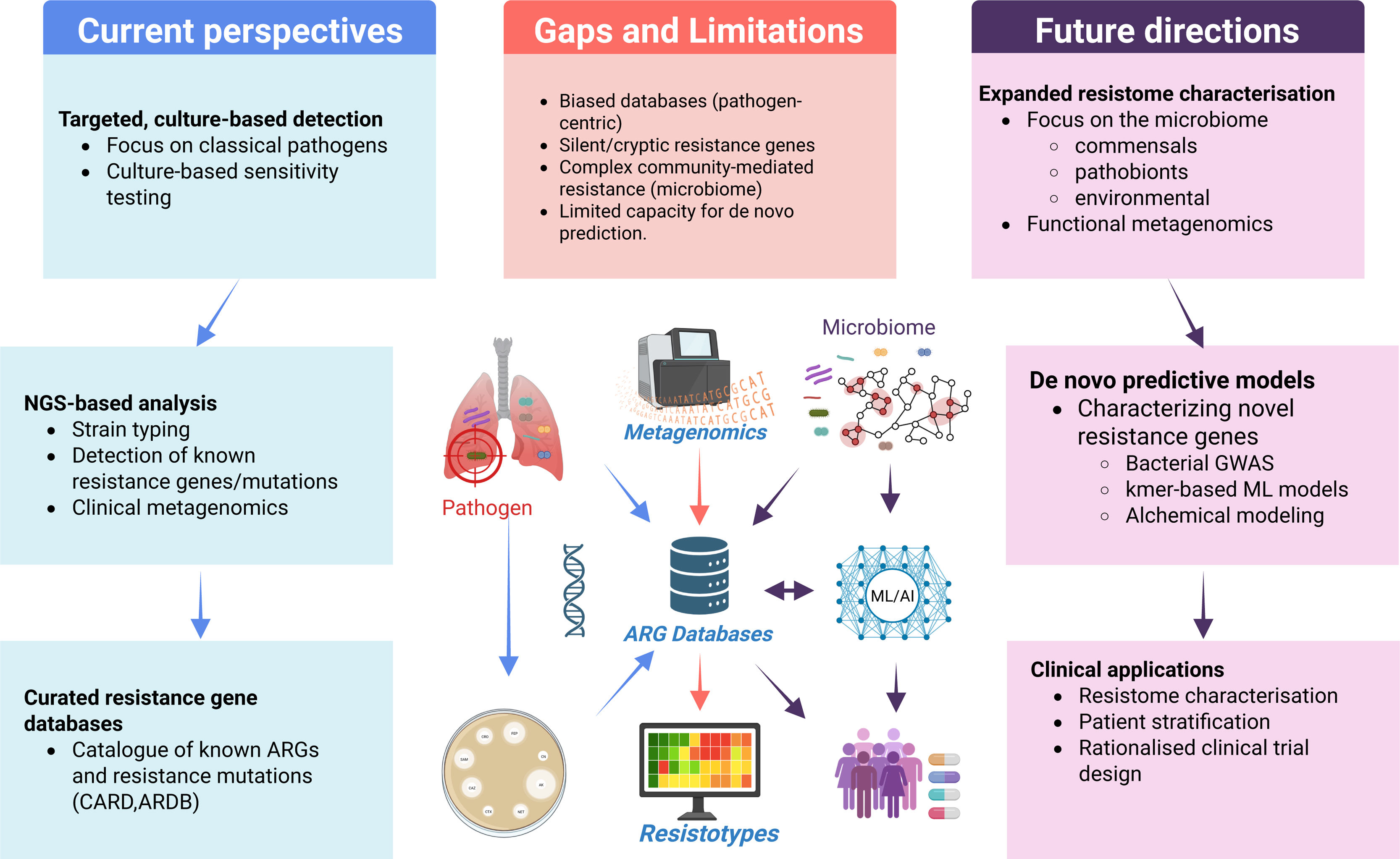
A conceptual framework for integrated resistome and microbiome models in clinical translation. Current models rely heavily on pathogen-centric ARG databases and culture-based methods focused on known resistance genes and organisms. These approaches are limited by database bias, inability to detect cryptic or uncharacterized resistance mechanisms, and poor resolution of complex community-level interactions. To overcome these gaps, expanded resistome profiling must incorporate commensals, pathobionts, and environmental microbes, leveraging metagenomics, functional analyses, and machine learning/artificial intelligence approaches (ML/AI) to iteratively supplement the ARG knowledge base. Future predictive frameworks combining bacterial GWAS, reference-free models, and de novo tools such as alchemical free energy modeling offer the potential to capture the full complexity of the airway resistome and guide precision therapy and trial design.
Microbiome studies increasingly highlight the critical role of community composition in bronchiectasis pathophysiology.2 Genera such as Neisseria, Rothia, and Aggregatibacter, previously unrecognized, have now emerged as important, with Neisseria linked to adverse outcomes and Rothia and Aggregatibacter exerting protective (anti-inflammatory) effects.3,13,14 Metagenomics reveal that antimicrobial resistance genes are widespread across pathogens, commensals and pathobionts, which collectively contribute to a “core” airway resistome, largely independent of disease status.9 Analysis of the Cohort of Asian and Matched European Bronchiectasis (CAMEB) has revealed distinct “resistotypes” correlating to clinical outcomes, where multidrug-dominant profiles link with frequent exacerbations and poorer lung function.15 Such associations have been independently validated in the European EMBARC-BRIDGE cohort demonstrating that, despite geographic variation in microbiomes, associations between key genera – pathogenic, commensal and pathobiont – including resistance profiles remains largely conserved.16 Taken together, this highlights the airway resistome as a critical determinant of microbiome resilience, therapeutic response and disease trajectory in bronchiectasis contrasting with the more traditional view that commensals represent passive bystanders.
Predicting antibiotic resistance from lung microbiomes is complicated by the presence of uncharacterized or ‘silent’ resistance genes activated through promoter mutations, mobile elements, or antibiotic selection.8,10 While extensively studied in pathogens, these processes remain largely unexplored in commensals. Here, it should be recognized that resistance mechanisms including CTX-M-type cephalosporinases, NDM-type metallo-β-lactamases and MCR-1-mediated colistin resistance all originated as uncharacterized genes within human and/or environmental bacterial communities, until their emergence in clinical pathogens in response to antibiotic selective pressures. Similarly, resistance mechanisms likely exist (undetected) in commensal and pathobiont constituents of the microbiome, silently influencing community dynamics under antibiotic pressure. This, in turn, may have protective or deleterious consequences depending on whether resistance promotes microbiome resilience against pathogenic invasion or facilitates an expansion of pathogenic species that disrupts beneficial microbial airway interactions.17,18 Current diagnostic frameworks rarely culture or examine commensal species, creating a “blind spot” in resistome surveillance.7 Consequently, microbiome models lack complexity, due to inherent biases in existing antimicrobial resistance gene (ARG) databases and the limited ability to predict uncharacterized resistance mechanisms de novo from metagenomic data alone. Enriching curated ARG databases with resistance genes from commensal and environmental microbes, combined with innovative machine learning and data-integrative approaches, will be critical in overcoming these limitations (Fig. 1). Here, metagenomics may offer a solution that guides therapeutic decision making allowing improved patient stratification. To achieve this shift necessitates pathogen-centric ARG databases to incorporate large-scale environmental and commensal-pathobiont WGS datasets coupled to predictive modeling which incorporates functional metagenomics, bacterial genome-wide association studies (GWAS), and pioneering tools such as alchemical free energy modeling to allow de novo prediction of novel resistance variants.19,20 Reference-free machine learning approaches, such as k-mer-based prediction offers an additional pathway to uncover novel resistance elements within the broader microbiome.21 Even if this is to be realized, significant computational challenges remain, including the need for scalable de novo ARG discovery pipelines and integrated frameworks capable of combining sequence, structure, and phenotype predictions into standardized clinically interpretable bioinformatic pipelines.7 Future models must address the confounding effects of horizontal gene transfer, genetic redundancy and complex community interactions, while striving to support predictive systems applicable to the broader microbiome as opposed to individual isolates.
As laboratory diagnostics and antibiotic sensitivity testing become increasing genome-centric and digitized, knowledge of the resistome must expand to improving clinical prediction. The resistome likely has a key role in understanding microbial survival under antibiotic pressure and may dictate how microbial ecosystems establish, persist, and respond to therapy within the airway. Clinical anomalies—such as the efficacy of azithromycin in treating P. aeruginosa exacerbations, the failure of targeted eradication therapies and inconsistent outcomes in bronchiectasis trials such as ORBIT and RESPIRE—suggest that the mechanistic underpinnings of therapeutic success (and failure) remain incompletely understood.22 Deeper integration of microbiome and resistome analyses will be key to advancing our understanding of the anti-microbial therapeutic response, guiding novel drug development and optimizing clinical trial design.22,23 Dynamic resistome profiling enables precision prescribing, early identification of high-risk microbiomes and offers ‘real-time’ prediction of treatment outcome with increased precision. By concurrently incorporating commensal resistomes into therapeutic planning, we recognize its dual role as a reservoir of resistance and potential mediator of microbial resilience. Machine learning algorithms, predictive molecular modeling and expanded environmental surveillance will further refine resistome interpretation beyond the constraints of curated clinical databases. Importantly, significant hurdles remain: defining comprehensive resistotypes, validating resistotype–phenotype associations and translating these into routine practice. Realizing the potential of the microbiome-resistome dynamic will necessitate moving beyond the narrow focus on classical pathogens, and embracing the complexity of microbial ecosystems, recognizing the resistome in all forms as a central determinant of clinical outcomes in bronchiectasis. Characterization of the resistome in bronchiectasis has significantly progressed in recent times illustrating that antibiotic resistance in chronic airways disease is a multifactorial phenomenon shaped by microbial ecology, host interaction and dynamic environmental pressure. Fully appreciating this will demand integrative models beyond genomic profiling and consideration of airway commensals and pathobionts, including their resistance mechanisms and survival strategies, as integrated components of the broader microbiome (Fig. 1).
Conflict of Interest StatementS.H.C. has served on advisory boards for CSL Behring, Pneumagen Ltd., Sanofi, Zaccha Pte. Ltd. and Boehringer Ingelheim, has served on Data Safety and Monitoring Boards (DSMB) for Inovio Pharmaceuticals Ltd and Imam Abdulrahman Bin Faisal University and received lecture fees from Astra-Zeneca and Chiesi Farmaceutici, all outside of the submitted work. All other authors have no conflicts to disclose.