Alpha-1 antitrypsin deficiency (AATD) is an autosomal co-dominant disorder that predisposes carriers to early development of chronic obstructive pulmonary disease (COPD) and/or liver disease. It is caused by the inheritance of 2 severe deficiency alleles in the SERPINA1 gene,1 and a plasma concentration of alpha-1 antitrypsin (AAT) below 50mg/dl is representative of a significant deficiency. Up to 95% of clinical cases related with AATD are associated with the PI*ZZ genotype, while the other 5% are associated with PI*SZ and PI*MZ genotypes or combinations of PI*S or PI*Z with other extremely rare deficiency or null alleles. These rare alleles account for 4.6% of the deleterious variants recorded in the Spanish Registry of Patients with AAT Deficiency, and null variants are very rare indeed.2 Although around 25 null variants have been discovered in the last 20 years or so, little information is available on their clinical impact.3–9 We report 2 cases of patients referred to the respiratory medicine clinic with a diagnosis of AATD associated with the PI*Q0amersfoort and PI*Q0cardiff allele.
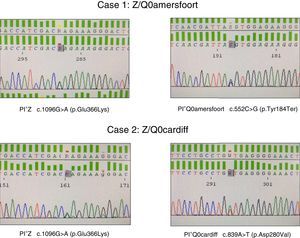
The first was a 47-year-old woman, native of the former republic of Yugoslavia, with a smoking history of 15 pack-years, referred to the respiratory medicine clinic for a 1-year history of dyspnea on moderate exertion (mMRC 2). Lung function tests results were as follows: FEV1/FVC 0.5; FEV1 1.80l (55%); FVC 3.40l (77%); DLCO 53%; KCO 52%. High-resolution computed tomography revealed centrilobular emphysema, predominantly in both lower lobes. Complete blood count, IgA, IgM, IgG, IgE and transaminases were within normal levels. Plasma AAT determined by nephelometry was 18mg/dl, so a genetic study to detect deficiency alleles S and Z was performed using real-time PCR (FRET, LightCycler 2.0, and TIB MOLBIOL probes), which detected heterozygosity for the PI*Z allele, while the presence of PI*S alleles was ruled out. In view of the discordance between the genotype obtained and AAT plasma levels, a molecular study was performed of all exonic coding regions and of the intronic sequences flanking the SERPINA1 gene, using Sanger sequencing (BigDye™ Terminator v3.1 Cycle Sequencing, Thermo Fisher Scientific). In this study, in addition to the PI*Z described above, heterozygosity for the PI*Q0amersfoort allele was detected (Fig. 1).
The second case was a 42-year-old man with no clinical history of interest, who was referred to our clinic for a family history of AATD. He had severe AAT deficiency (41mg/dl), and heterozygosity for the PI*Z allele was detected. Molecular study of the SERPINA1 gene revealed a Z/Q0cardiff genotype (Fig. 1). The patient had no respiratory symptoms or liver disease. Lung function test results were within normal limits: FEV1/FVC 0.79; FEV1 4.82l (109%); FVC 5.60l (112%); DLCO 109%; KCO 106%. Chest radiograph and general laboratory tests were unremarkable.
AAT is an antiprotease, produced mainly by the hepatocytes, which inhibits the elastase activity of the neutrophils. Normal plasma levels range between 120 and 200mg/dl.1 Although the Z mutation (p.Glu 342 Lys) deficiency allele is the most common and leads to very low AAT levels in plasma (around 10%–15% of the normal level), serum AAT levels in null mutations are extremely low or undetectable3–9: a wide range of molecular mechanisms are involved in this outcome, including errors in protein synthesis or post-translational degradation.6,10–12 For this reason, genotypes consisting of null homozygotes or accompanied by other deficiency alleles of the SERPINA1 gene carry a particularly high risk of very early onset pulmonary emphysema, even earlier than would be expected in the ZZ genotype.13
With regard to PI*Q0amersfoort, the limited literature available suggests that both heterozygous and homozygous forms lead to COPD at an early age, as observed in our female patient, with no liver involvement whatsoever.7,8 This mutation causes a stop codon at position 184 of the protein, resulting in a severe deficiency when associated with other deficiency variants, such as PI*Z. Like other null alleles, the PI*Q0amersfoort variant does not cause liver disease because the protein is not polymerized in the liver, as occurs in mutations caused by amino acid switches, in which deficiencies exist that alter protein structure and folding, giving rise to accumulation in the endoplasmic reticulum of the hepatocytes, resulting ultimately in tissue damage.
In the case of PI*Q0cardiff, the amino acid aspartate is replaced by valine at position 256 of the ATT protein. This substitution causes a severe deficiency when it occurs in homozygosis or in association with other deficiency variants such as PI*Z. Some authors argue that homozygous Q0cardiff patients are not at risk for emphysema, although there may be a risk if it occurs in heterozygosity with the PI*Z allele or other null alleles.9,14 In our case, the male patient was asymptomatic. In our opinion, PI*Q0cardiff cannot be considered a null allele, since it is not a mutation that causes a premature stop codon, nor does it produce complete protein degradation, a mechanism usually observed in null deficiency alleles.5 Some authors define the PI*Q0cardiff variant as PI*P(lowell), since the genetic variation consisting of the switch of an aspartic acid to valine9 causes degradation of the intracellular protein, which leads to reduced, but not undetectable, levels of protein.15 It seems likely that these residual protein levels, along with those produced by the PI*Z allele, caused our patient to present AAT levels higher than those found in patients with null alleles.
Please cite this article as: Figueira Gonçalves JM, Martínez Bugallo F, García-Talavera I, Rodríguez González J. Déficit de alfa-1-antitripsina asociado a alelos nulos. Arch Bronconeumol. 2017;53:700–702.