In this narrative review, we address the ongoing challenges of lung cancer (LC) screening using chest low-dose computerized tomography (LDCT) and explore the contributions of artificial intelligence (AI), in overcoming them. We focus on evaluating the initial (baseline) LDCT examination, which provides a wealth of information relevant to the screening participant's health. This includes the detection of large-size prevalent LC and small-size malignant nodules that are typically diagnosed as LCs upon growth in subsequent annual LDCT scans. Additionally, the baseline LDCT examination provides valuable information about smoking-related comorbidities, including cardiovascular disease, chronic obstructive pulmonary disease, and interstitial lung disease (ILD), by identifying relevant markers. Notably, these comorbidities, despite the slow progression of their markers, collectively exceed LC as ultimate causes of death at follow-up in LC screening participants. Computer-assisted diagnosis tools currently improve the reproducibility of radiologic readings and reduce the false negative rate of LDCT. Deep learning (DL) tools that analyze the radiomic features of lung nodules are being developed to distinguish between benign and malignant nodules. Furthermore, AI tools can predict the risk of LC in the years following a baseline LDCT. AI tools that analyze baseline LDCT examinations can also compute the risk of cardiovascular disease or death, paving the way for personalized screening interventions. Additionally, DL tools are available for assessing osteoporosis and ILD, which helps refine the individual's current and future health profile. The primary obstacles to AI integration into the LDCT screening pathway are the generalizability of performance and the explainability.
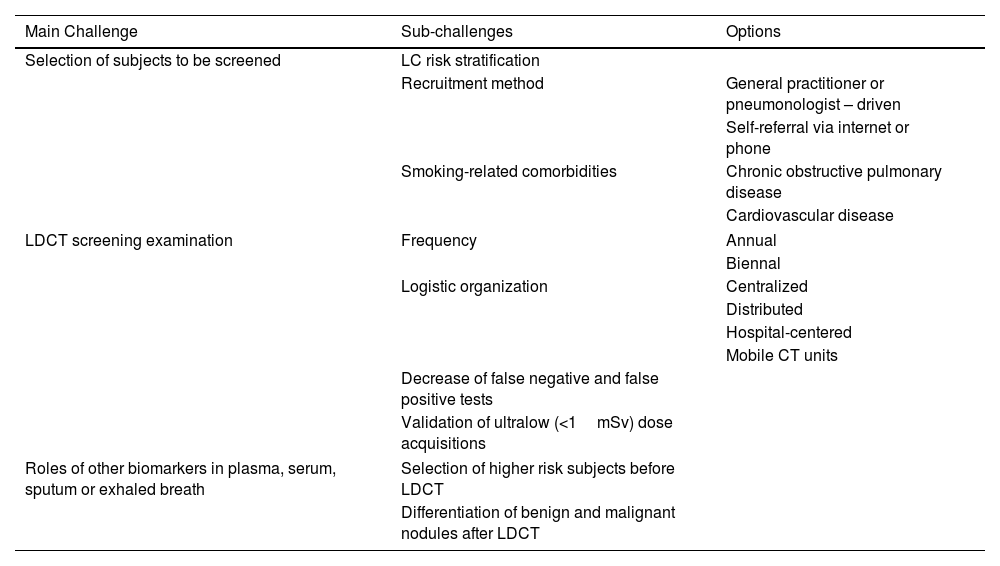
Lung cancer (LC) is the leading cause of cancer-related deaths worldwide.1 While smoking and age are the primary risk factors for LC, making smoking cessation the main preventive measure, two randomized clinical trials – the National Lung Screening Trial (NLST)2 in the US and the NELSON3 in Europe – have demonstrated that annual screening with low-dose computed tomography (LDCT) significantly reduces mortality from LC compared to annual chest X-rays or no screening. Consequently, LC screening with annual LDCT is recommended for smokers or former smokers aged 50–80 years.4,5 However, the reduction in LC mortality associated with LDCT screening is modest. A meta-analysis of nine trials reported an average relative risk of 0.84 for LC mortality (95% CI: 0.76–0.92) in LDCT-screened subjects compared to non-screened subjects.6 This justifies efforts to enhance LC screening with LDCT by addressing its persistent challenges7,8 in selecting subjects for screening,9–13 improving the LDCT screening examination,3,10–18 and incorporating other biomarkers from plasma, serum, sputum, or exhaled breath (Table 1).11–17
Challenges of LC Screening With LDCT.
| Main Challenge | Sub-challenges | Options |
|---|---|---|
| Selection of subjects to be screened | LC risk stratification | |
| Recruitment method | General practitioner or pneumonologist – driven | |
| Self-referral via internet or phone | ||
| Smoking-related comorbidities | Chronic obstructive pulmonary disease | |
| Cardiovascular disease | ||
| LDCT screening examination | Frequency | Annual |
| Biennal | ||
| Logistic organization | Centralized | |
| Distributed | ||
| Hospital-centered | ||
| Mobile CT units | ||
| Decrease of false negative and false positive tests | ||
| Validation of ultralow (<1mSv) dose acquisitions | ||
| Roles of other biomarkers in plasma, serum, sputum or exhaled breath | Selection of higher risk subjects before LDCT | |
| Differentiation of benign and malignant nodules after LDCT | ||
This article aims to review the established achievements and ongoing efforts in addressing some challenges of LC screening through artificial intelligence (AI) applications. Specifically, we focus on AI tools that evaluate the baseline LDCT, which is the most crucial examination in the LC screening regimen from an individual health perspective.
The Pivotal Role of Baseline LDCT for LC ScreeningParticipants in LC screening programs typically undergo annual LDCT examinations and, if abnormalities are found, further tests to diagnose or exclude LC. The baseline (first) LDCT is crucial for several reasons. First, most LCs diagnosed during the initial 2–4 annual screening rounds are already present in the baseline LDCT. In particular, screen-detected LCs diagnosed within the first year following the initial LDCT screening test, defined as prevalent LCs (Fig. 1), are typically more numerous (range 55.4–84%) than those diagnosed after the subsequent annual repeat LDCT screening, defined as incident LCs.2,3,18–22 Moreover, most (77–80%) incident LCs are already present in baseline (or prior) LDCT scans20,23 (Fig. 1). However, these “pseudo-incidental” LCs require time to grow and reach a size threshold that qualifies them as suspicious or actionable nodules, and can ultimately be diagnosed as LCs only years after their appearance. The combination of prevalent and “pseudo-incidental” LCs allows the retrospective identification of malignant lesions in the baseline LDCT in up to 92% of subjects with screen-detected LCs within the first three-four years of screening.13,20,24 Awareness of the distribution of screen-detected LCs is essential given the expected new start and adoption of LC screening as a population-based intervention in Europe4 and elsewhere. Second, baseline LDCT allows the extraction of markers of smoking-related comorbidities, such as coronary artery calcifications (CAC) for cardiovascular disease (CVD) and pulmonary emphysema for chronic obstructive pulmonary disease (COPD). In particular, pulmonary emphysema can be assessed using visual semi-quantitative scales25,26 (Figs. 2 and 3) or quantitatively with the extraction of several indices using automatic software, including deep learning (DL) algorithms27–30 (see “CVD, Respiratory and Overall Mortality Prediction” section). Emphysema is associated with an increased LC incidence,31–33 but, more importantly, in the perspective of LC screening programs, both CAC and emphysema indices predict long-term overall, CVD and respiratory mortality25,34–37 (Figs. 2 and 3). For this reason, in principle, LDCT assessment of CAC and emphysema allows for screening regimen personalization9 and early initiation of therapies that can delay comorbidities progression. A compelling argument underscoring the pivotal role of LDCT is that the assessment of smoking-related disease markers, such as CAC and emphysema indices, in the baseline LDCT provides sufficient prognostic information at the individual level. In fact, longitudinal studies have shown that only about 15% of subjects with emphysema, who participated in LC screening, experienced a mild progression of emphysema itself.38 Also the progression of CAC is relatively slow, with only one out of five subjects without CAC developing some within 4–5 years.39 Third, changes consistent with interstitial lung abnormalities (ILA) or disease (ILD) are observed in 3–10% of subjects undergoing baseline LDCT40 (Fig. 5). These changes imply a greater risk of LC and are associated with an increased rate of complications from LC treatments.41 Detection of these abnormalities, especially when they extend to at least 5% of the lung parenchyma, justifies referral to a multidisciplinary team to prevent and manage their progression40,42 (Fig. 5). Fourth, the baseline LDCT can reveal several additional incidental findings, the most important and frequent being bronchiectasis, consolidations, aortic valve disease, mediastinal masses, enlarged mediastinal or hilar lymph nodes, and thyroid abnormalities.40 Fifth, eligible subjects often undergo baseline LDCT only and then quit the screening program. In fact, in the US, adherence to the recommended screening intervals can be as low as 57%, especially among subjects with negative tests or benign nodules.43 Finally, in the UK LC Screening trial, which offered just one LDCT to eligible subjects of the intervention arm,44 a decrease in mortality from LC was observed in the screened subjects compared to controls (no screening).6 This benefit might be valuable for deprived world areas where limited economic resources do not allow serial annual LDCT examinations.
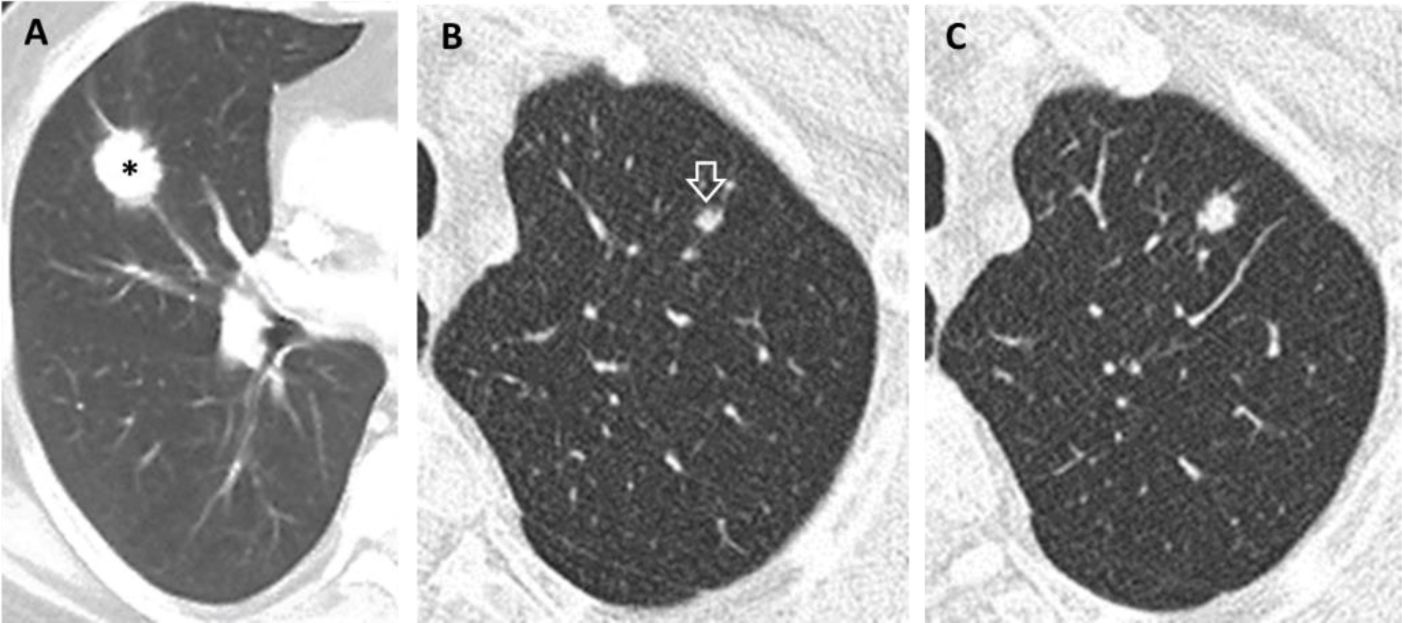
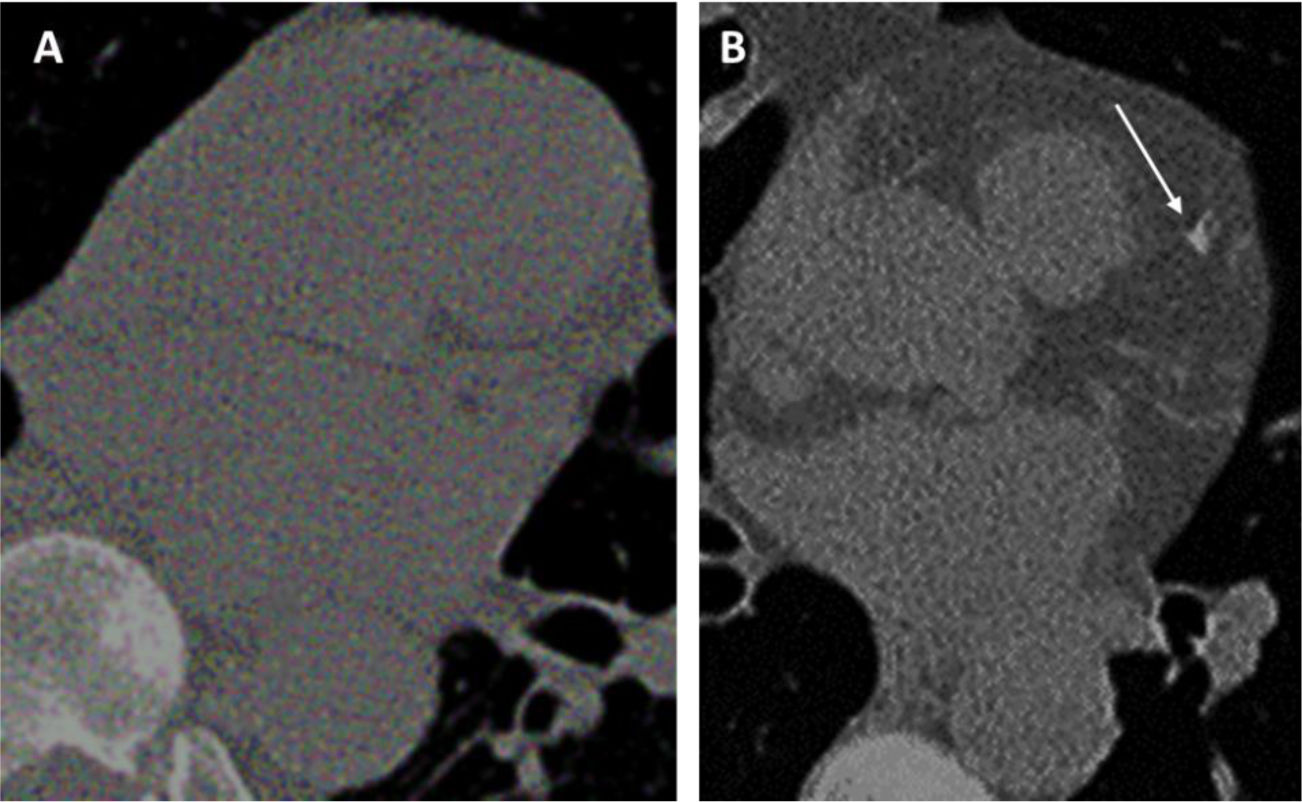
(A–C). Prevalent and pseudo-incidental screen-detected LC at baseline LDCT. Stage IA adenocarcinoma in a 60-year-old man from ITALUNG (A) appearing at baseline LDCT as a large (26mm in mean diameter) solid nodule in the right upper lobe (*). Pseudo-incidental stage IA squamous cell carcinoma in a 67-year-old man from ITALUNG (B and C) appearing at baseline LDCT (B) as an infra-threshold (5.2mm in mean diameter) solid nodule in the left anterior lobe (white empty arrowhead) and showing growth (10mm in mean diameter) at the first annual repeat (C).
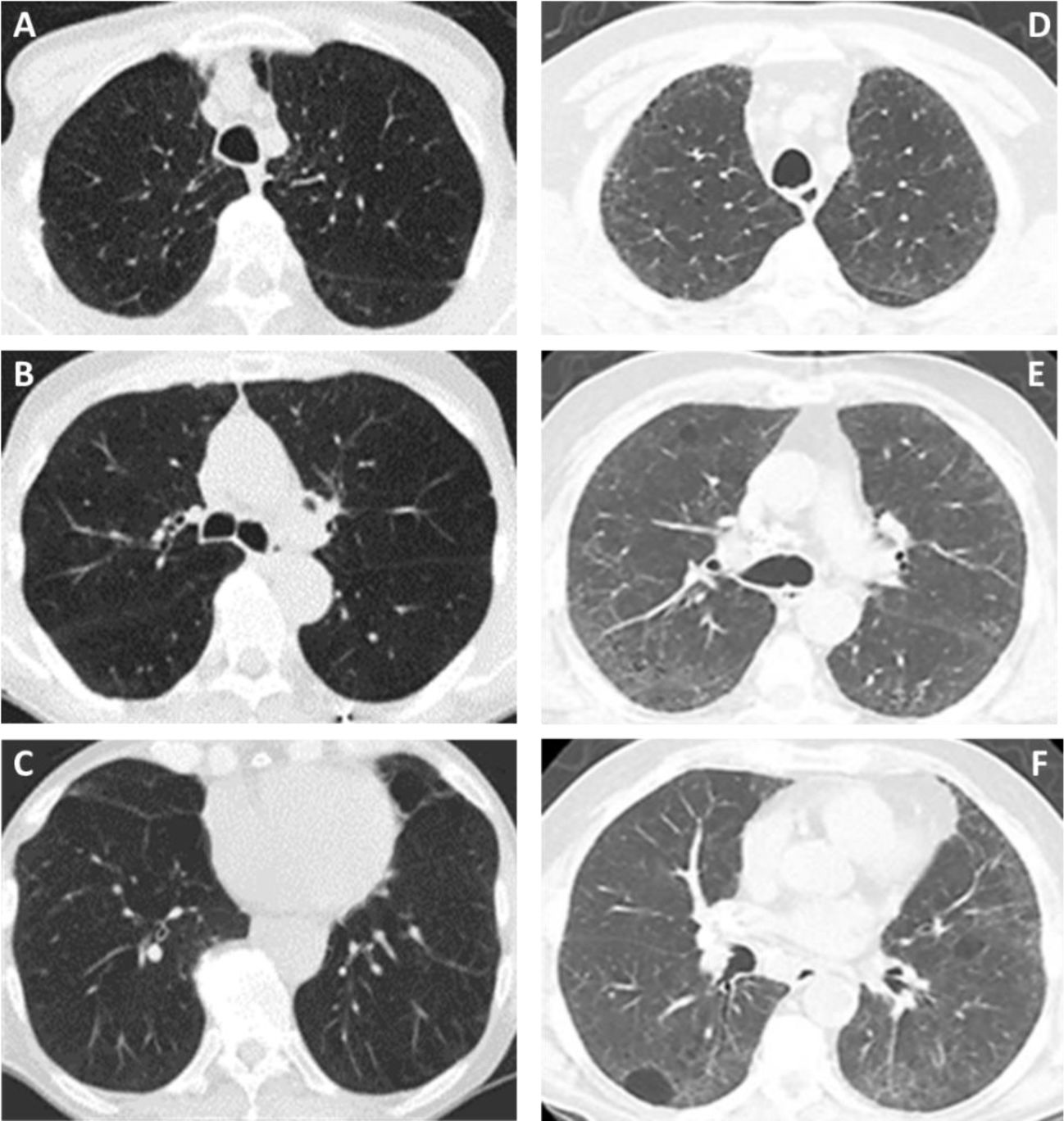
(A–F) Diffuse lung disease at baseline LDCT. Advanced destructive pulmonary emphysema (A–C) in a 65-year-old man from NLST who died of respiratory disease (ICD code J449) 835 days after randomization. Interstitial lung disease (D–F) in a 73-year-old man from NLST who died of respiratory disease (ICD code J849 – interstitial pulmonary disease unspecified) 2462 days after randomization.
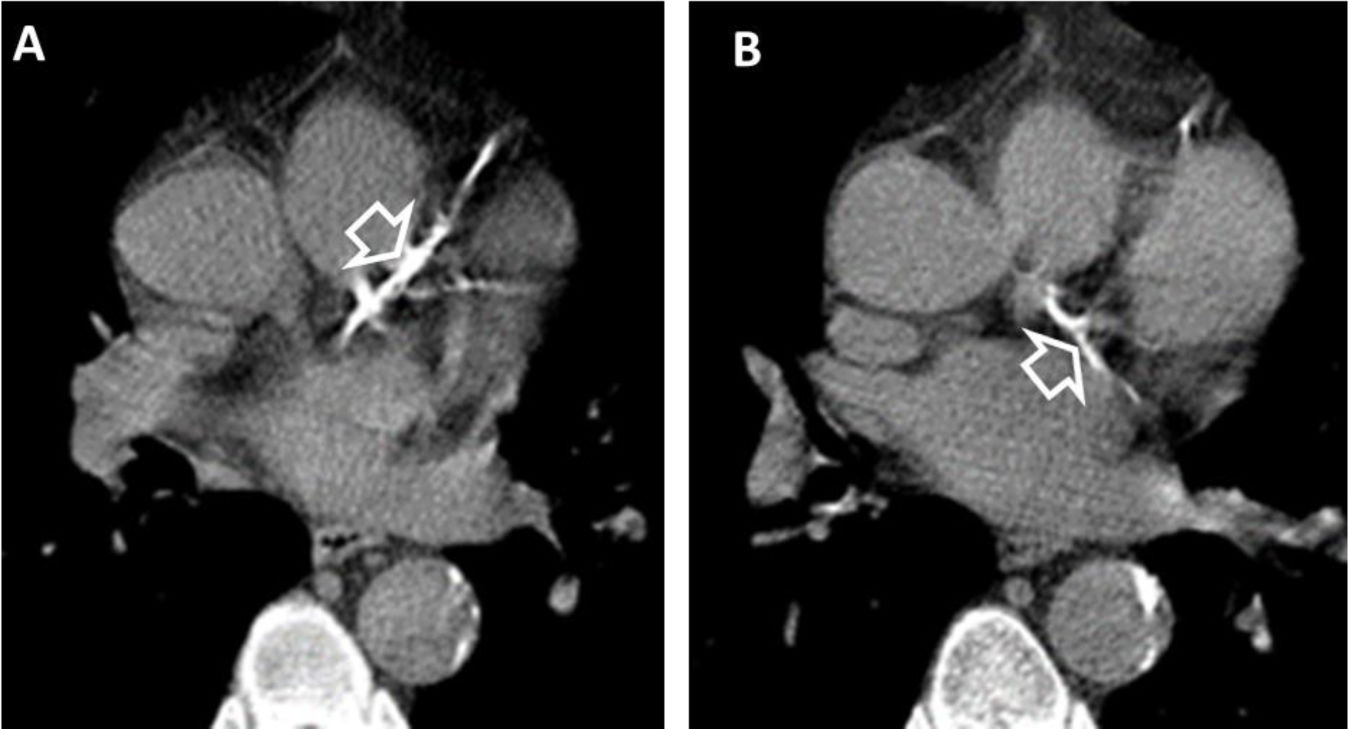
(A and B). Coronary artery calcifications at baseline LDCT. Severe coronary artery calcifications in the anterior interventricular artery (white empty arrowhead A) and left circumflex artery (white empty arrowhead B) at baseline LDCT in a 69-year-old man from NLST who died of atherosclerotic heart disease (ICD code I251) 226 days after randomization.
In the usual screening workflow, each LDCT examination undergoes a double reading by radiologists, who meticulously examine them for early signs of cancer,45 focusing on the characteristics of the pulmonary nodules, including size, morphology, location, and change over time. The LDCT examination also allows the opportunistic assessment of smoking-related comorbidities, especially emphysema and CVD.46 This makes medical image interpretation the cornerstone of LC screening activities, requiring significant time and expertise.47 With the new USPSTF guidelines expanding the cohort of eligible individuals for LC screening in the US,48 the already high radiologists workload49 is expected to increase further, making fully manual reporting of LDCT examinations impractical.
In recent years, the integration of AI into healthcare has brought significant changes in LC screening practice. By leveraging machine learning (ML) and DL algorithms (see Yu et al.50 for a review), researchers and clinicians can efficiently harness the vast amounts of data generated by LDCT to address critical challenges in LC screening. This section explores diverse applications of AI in baseline LDCT imaging for problem-solving in LC screening.
CAD for Lung NodulesDetecting lung nodules in LDCT images is central to LC screening workflows, as it guides participant management. However, the repetitive nature of this task and the overwhelming volume of images contribute to high intra- and inter-observer variability and a high false positive rate.51,52 Computer-aided diagnosis (CAD) systems assist radiologists by automatically identifying subtle findings, thereby mitigating human limitations like memory, distraction, and fatigue and offering objective data interpretation.53 Computer-aided detection (CADe) systems are used for detection, while computer-aided diagnosis (CADx) systems are used for diagnosis.51,53 CADe systems have been shown to reduce the rate of false-negative baseline LDCT examinations.3,54–61 Additionally, they can help detect infra-threshold nodules that do not qualify as positive according to Lung-RADS,62 but need to be monitored in subsequent LDCT examinations.29 However, only a small fraction (below 1%) of micronodules (<4mm) evolves into LC,63 indicating that the specificity of a micronodule at baseline LDCT is extremely low.
Using CADe for the computation of lung nodule volume rather than diameters has improved classification of nodules and decreased the number of indeterminate or false positive LDCT examinations.64 However, the clinical integration of CADe remains limited due to persistent concerns over high false positive rates.65,66 Researchers are addressing this issue through several strategies. CADe tools may be used as pre-screening instruments to rule out negative LDCT examinations, allowing radiologists to concentrate on more challenging and suspicious cases.67,68 Another strategy involves integrating more data into the models. For example, a ‘collaborative CAD’ system incorporating radiologists’ gaze patterns into a 3D multi-task convolutional neural network (CNN), a particular DL architecture,69 achieved a 97% classification accuracy in identifying nodules.70
In LC screening, it is crucial to distinguish between benign lung nodules, which constitute the vast majority observed in low-dose CT scans of screened subjects according to Lung-RADS v2022,62 and malignant lung nodules. This differentiation often leads to additional examinations, such as follow-up LDCTs at intervals of 1–3–6 months, FDG-PET,71 and invasive procedures, significantly increasing both the costs and potential harms associated with screening.7 Notably, malignant nodules demonstrate an increase in size, density, or both over subsequent 3 or 6-month follow-up LDCT scans, as outlined in Lung-RADS v2022.62 The calculation of volume doubling time (VDT) serves as a practical and effective method to assess nodule growth characteristics and malignancy risk.64 The Lung-RADS guidelines recommend specific management strategies for baseline LDCT-detected nodules, particularly solid non-calcified nodules ≥6mm in diameter or ≥113mm3 in volume, which helps streamline further investigations aimed at confirming malignancy and minimizing unnecessary procedures.72 Furthermore, this differentiation can be enhanced by integrating LDCT features such as nodule size and density, the number of nodules, and presence of emphysema, with pertinent subject history, as incorporated in the PanCan/Brock model,73,74 or with biomarker results such as plasma DNA methylation75 or plasma total cfDNA.13 However, the PanCan/Brock model has been developed, tested, and calibrated specifically for prevalent solid nodules ≥6mm in diameter.73,74,76 It may not be well-suited for newly appearing nodules detected at next LDCT screening rounds.76 Additionally, this model may not effectively identify malignant micronodules, potentially leading to the delayed (“pseudo-incidental”) LC diagnosis. Therefore, DL algorithms predicting LC based on baseline LDCT and radiomics77–80 may improve the characterization of these small nodules. For example, an ML approach combining epidemiological, clinical and radiomic features, extracted from the nodules present at baseline LDCT, was able to predict the nodule's malignancy risk score with an area under receiving operator curve (AUROC) of 0.93, outperforming the PanCan/Brock model and with optimal performance for both solid and sub-solid nodules.81 Still, a generative approach to enhance the characterization of indeterminate nodules from the baseline LDCT scan80 exploited a growth model based on the Wasserstein generative adversarial network framework (GP-WGAN) to predict the nodule growth patterns in the 1-year follow-up LDCT scans. By leveraging the ability of GANs to generate data similar to the original, they can simulate follow-up LDCT examinations requiring only the baseline LDCT as input. The results demonstrated that the generated follow-up nodule images, when used as input to a model for LC malignancy prediction, achieved performance comparable to using real follow-up nodule images (AUROC of 0.82±0.02 for generated nodules, compared to 0.86±0.02 for real nodules).80
LC Risk StratificationFor LC screening to be effective and minimize related harms, it is crucial to carefully select the at-risk population.49 Once selected, LDCT examination information allows for valuable risk stratification, enabling a tailored screening schedule.82 Several models for estimating LC risk have incorporated baseline LDCT findings.83–85 Their implementation is hindered by limited external validation and the need for manual input of LDCT findings into the model to calculate the score. Unlike traditional models, AI-based algorithms autonomously analyze the entire LDCT volume, identify lung nodules and incidental findings, and combine this information with demographic data to generate a comprehensive, automated risk score.
The Google DL model evaluates LDCT examinations to predict LC incidence. It extracts local and global features from the current, and optionally prior, LDCT examinations and estimates the likelihood of a LC diagnosis within a year.86 Despite achieving a high AUROC of 0.959 on single LDCT examination and outperforming radiologists, the model was criticized for its ‘black-box’ nature, lack of source code availability, and small validation set.87
DeepScreener is a DL algorithm designed to predict a patient's cancer status from CT scans through three tasks: nodule segmentation, nodule-level classification and patient-level classification.88,89 For each nodule, the nodule-level classifier extracts morphological, textural and other features and combines them with the nodule location to calculate a risk score. Subsequently, the patient-level classifier aggregates the risk scores of all detected nodules to generate an overall risk score for the patient and determine the label (“cancer” or “no cancer”). The model achieved an AUROC of 0.89 and a sensitivity of only 42.4%, indicating that further refinement and validation are needed.89
Sybil, a DL model designed to predict using a 0–1 score the LC risk from a single LDCT examination up to the next six years, without the need for radiologist annotations or additional data, represents a recent advancement.90 It achieved AUROC for LC prediction at one year of 0.86–0.94 in three different datasets. Interestingly, when Sybil predicts high LC risk, the used signal localizes to specific at-risk regions rather than being equally spread over the entire thorax.90
DL algorithms such as Sybil could be used to stratify the risk of LC after a baseline LDCT and could be particularly valuable in providing the LC risk in a screened subject showing infra-threshold nodules, that correspond to benign or pseudo-incident LC, or, after a negative baseline LDCT, anticipating interval or incident LC. Examples of application of the Sybil algorithm are shown in Fig. 4.
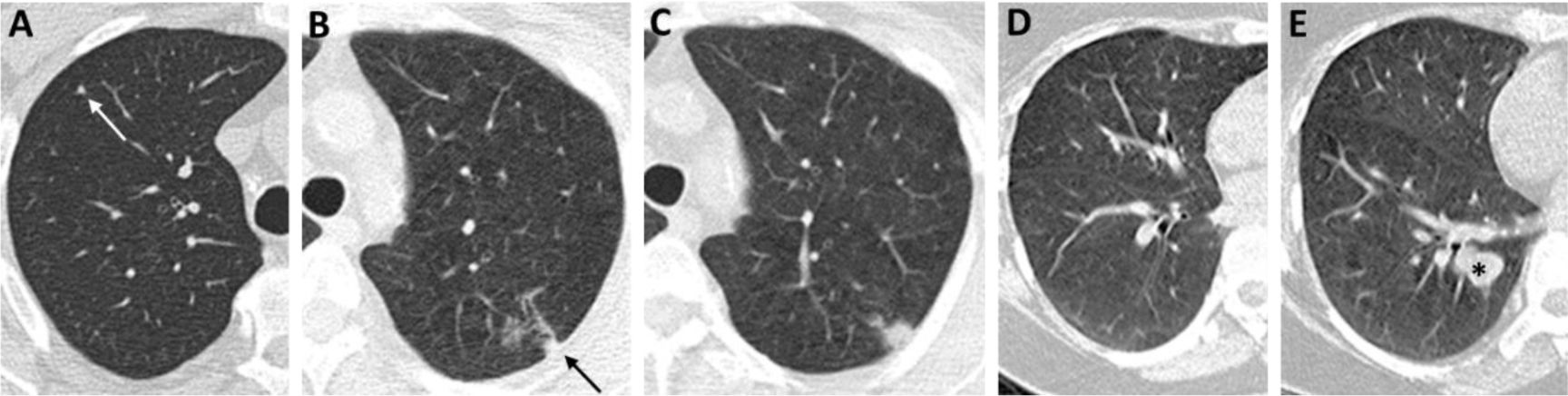
(A–E) Assessment of risk of LC in the next 1–6 years based on the analysis of baseline LDCT with the Sybil deep learning algorithm.90 (A) Prediction of a very low probability of LC after 1 year (risk score=0.0109) and 6 years (risk score=0.0831) since baseline LDCT in a 59-year-old man from NLST with a small infra-threshold (1.8mm in mean diameter) solid benign nodule in the right upper lobe (white arrow) at baseline LDCT who was alive 11 years after randomization. (B and C) Prediction of a moderate probability of LC after 1 year (risk score=0.3057) and 6 years (risk score=0.5998) since baseline LDCT in a 56-year-old woman from NLST with a small infra-threshold (3.8mm in mean diameter) (black arrow) solid nodule in the left upper lobe at baseline LDCT (B) which showed growth (9mm in mean diameter) at the annual LDCT performed two years later (C) consistent with a pseudo-incidental LC and who received a diagnosis of stage IA adenocarcinoma and was alive 11 years after randomization. (D and E) Prediction of a very low risk of LC after 1 (risk score=0.0017) and 6 years (risk score=0.0329) since baseline LDCT in a 57-year-old woman from NLST with a negative baseline LDCT (D) who showed a large (18mm in mean diameter) solid lesion (*) at the next annual LDCT (E) consistent with an incident LC who received a diagnosis of small cell carcinoma and died of LC (ICD code C349) 1559 days after randomization.
Tobacco smoking is a well-established risk factor for CVD, COPD, and LC. In LC screening cohorts, which primarily include current and former smokers, these conditions are the leading causes of death,2,3,91,92 and are often referred as the ‘Big 3 killers’. Using AI to extract comorbidity-related biomarkers from baseline LDCT images offers a valuable opportunity to enhance LC screening. AI can help optimize screening schedules – such as determining when to start, how frequently to screen, and when to stop – by refining individual risk profiles.9 Although radiologists’ visual scoring of comorbidities provides adequate predictive values25,35 (Figs. 2 and 3), AI-derived biomarkers offer greater robustness and objectivity, all without increasing the clinician's workload.93 One significant proof of this concept is a DL algorithm for the automatic quantification of coronary calcium.94 The resulting calcium scoring showed a high correlation with readings from expert radiologists and demonstrated robust test-retest accuracy.94 Beyond using AI-derived CAC as a predictor of CV events in LC screening cohorts,94–97 researchers are exploring additional approaches. For instance, a model was developed that based on the extraction of the coronary calcium and juxta-cardiac fat uses a single LDCT examination and provides a 0–1 score to estimate the probability of CVD risk.98 The model's ability to predict the risk of CVD and CV mortality equalized or surpassed that of radiologists and surpassed that of other state-of-the-art DL tools.94,98,99 Examples of its application to predict CV death in subjects with no or mild CAC are shown in Fig. 5. Other DL-derived indices include the prediction of adverse events based on the left atrial volume100 and of CV risk based on epicardial adipose tissue amount alone.101
(A and B) Assessment of risk of CV disease based on the analysis of baseline LDCT with Chao et al. deep learning algorithm.98 The algorithm attributes a moderate (score=0.351) CV risk in a 55-year-old man from NLST who did not show any coronary artery calcification at baseline LDCT (A) and who died of ischemic heart disease (ICD code I250) 2004 days after randomization. The algorithm attributes a high (score=0.700) CV risk in a 70-year-old woman from NLST with mild coronary artery calcifications (white arrow) at baseline LDCT (C) and who died of acute myocardial infarct (ICD code I219) 511 days after randomization.
COPD is typically diagnosed and evaluated through symptom assessment, spirometric testing, and tracking respiratory exacerbations.102 While lung densitometry is more reproducible than visual assessment of emphysema103 and is increasingly used for COPD assessment,29,31,104 it is notably sensitive to variations in CT scanners and acquisition/reconstruction parameters, such as slice thickness, radiation dose, and reconstruction kernel.105 A two-step DL model was developed to normalize the kernel effect for emphysema quantification in LDCT Images.105,106 This tool allows accurate emphysema quantification even when images are reconstructed using different kernels, thus improving consistency across large screening trials.105
Combining quantitative and semi-quantitative biomarkers for CVD and COPD in risk stratification after LDCT examinations is gaining attention. A logistic regression model that integrates participant demographics with LDCT measures of LC, CVD and COPD was developed to predict the 5-year risk of competing death. This approach helps identify individuals who may benefit more from preventive care for other conditions than from LC screening.107 The results suggest that a model based exclusively on quantitative LDCT measures, even when automatically derived, is suitable for calculating risk scores in a LC screening cohort and informing the post-LDCT screening process. Similarly, the predictive value of CAC visual score and of densitometry assessment of emphysema (relative area of the lung with density below −950 Hounsfield Units – RA950) in baseline LDCT along with age, gender, smoking status and pack-years were evaluated to predict the overall, LC, and CVD mortality in a screening cohort.36 Using an ML paradigm based on decision trees108 and the SHAP framework109 to assess the importance of each feature, the model interpretation revealed that RA950 was the first ranking feature for predicting overall and CVD mortality, with AUROC values of 0.70 and 0.73, respectively. The most important features for predicting LC mortality were pack-years and RA950, with an AUROC of 0.61.
Osteoporosis AssessmentCOPD is frequently associated with other extra-pulmonary systemic manifestations, including osteoporosis,110 that leads to an increased risk of fractures.111 Since bone attenuation measured on routine chest CT has shown strong correlation with bone mass density (BMD) assessed by dual-energy X-ray absorptiometry (DXA) in patients with COPD,112 opportunistic DL-aided assessment of osteoporosis in LDCT scans in LC screening cohorts has emerged.
Different approaches have been proposed. A DL model was combined with geometric operations to automatically measure BMD from LDCT scans achieving a good agreement with quantitative CT.113 AI-RAD Companion was evaluated as an end-to-end solution to derive a LDCT biomarker for osteoporosis in LC screening whose score moderately correlated with WHO T-scores allowing to stratify participants into normal, osteopenia, and osteoporosis categories.114 Additionally, the combination of ML with radiomics texture analysis of automatically detected vertebral body achieved an AUROC of 0.90 and 0.72 on internal and external validation cohorts, respectively,115 establishing that osteoporosis can be part of the evaluation of LDCT for LC screening with impacts on morbidity, mortality, and the overall efficacy of LC screening.
Classification and Prediction of ILD EvolutionRecently, several studies have demonstrated the capability of DL algorithms to help classify the ILD detected in full dose thin-section CT116–119 and, more importantly, to predict the progression of the disease and the mortality due to this condition.120–122 Validation of these algorithms in the LDCT examinations performed for LC screening is still required.
ConclusionsWhile numerous AI models have been developed for LC screening, significant challenges remain that hinder their effective integration into clinical practice. Key issues include the generalizability of AI models across different populations – complicated by the limited availability of open-access datasets –, the explainability of AI decisions,47,52,65,123,124 and the assessment of AI tools deployment. These concerns have been extensively discussed in recent literature,125–129 highlighting the urgent need for ongoing research and collaboration in this rapidly evolving field.
The authors state that they have no conflict of interests.
The authors thank the National Cancer Institute for access to NCI's data collected by the National Lung Screening Trial (NLST) – CDAS Project Number: NLST-1175.
The research leading to these results has received funding from the European Union – NextGenerationEU through the Italian Ministry of University and Research under PNRR – M4C2-I1.3 Project PE_00000019 “HEAL ITALIA” to Stefano Diciotti – CUP J33C22002920006. The views and opinions expressed are those of the authors only and do not necessarily reflect those of the European Union or the European Commission. Neither the European Union nor the European Commission can be held responsible for them.