Post-COVID syndrome substantially affects the patient's quality of life, yet diagnosis is often difficult.1 Therefore, there is an unmet need for developing and validating novel biomarkers that will not only help to establish the diagnosis but also enable defining treatment response. Although both CT-based and X-ray-based radiomic studies successfully distinguish COVID-19 pneumonia from other types of pneumonia and allow the prognostic evaluation of the disease,2,3 there are no evident radiomic features to date that are predictive of the development of long-term symptoms. Therefore, in this work, we aimed to evaluate the role of deep learning-boosted X-ray radiomics in diagnosing post-COVID patients.
The study population consisted of 260, 278, and 435 COVID-19 (mild to moderate acute disease), post-COVID and non-COVID patients, respectively. According to the World Health Organization's (WHO) definition, patients who continued to have or developed new symptoms three months after the initial SARS-CoV-2 infection (with these symptoms lasting for at least two months with no other explanation) were considered individuals with post-COVID syndrome.4 These patients mostly presented with respiratory symptoms (e.g. persistent dyspnea and/or coughing); however, individuals with cardiovascular (e.g. palpitation) manifestations were also included. Post-COVID individuals were diagnosed and examined at the Institution's multidisciplinary post-COVID department. Of note, upon enrollment, all patients were required to complete a standardized questionnaire that covered various aspects of previous infections and symptoms, and they underwent clinical testing to confirm the diagnosis of post-COVID syndrome. As for the COVID-19 patients, inclusion criteria consisted of a confirmed diagnosis of acute SARS-CoV-2 infection by PCR assay requiring hospitalization. Post-COVID and COVID-19 patients with a recent history of intensive care unit admission were excluded. Non-COVID patients were referred from our institution's outpatient department. These patients were diagnosed with mild forms of chronic lung disease, including chronic obstructive pulmonary disease or asthma, and their chest X-rays (CXRs) were performed as part of the routine check-up in the pre-pandemic era (at least six months before the COVID-19 outbreak). All aspects of the current study were conducted in strict compliance with the ethical standards and principles outlined in the Declaration of Helsinki. The study protocol was approved by the national-level Ethics Committee (3270-3/2022/EÜIG).
Radiomic workflow began with lung segmentation and cardiothoracic ratio (CTR) determination. For the initial steps of algorithm development for lung segmentation we used the Japanese Society of Radiological Technology's (JSRT) CXR dataset, with pre-divided training and test sets.5 This dataset is particularly valuable for training and/or externally validating radiomics algorithms based on CXRs, as it contains a substantial collection of digitized PA CXRs gathered from 14 medical centers, all reviewed and annotated by board-certified radiologists. After the neural network was trained to learn the segmentation of the two lungs separately, the segmentation of the heart, and the background in each layer (resulting in a 4-layer segmentation of the same size as the input), we utilized a binary morphological closure algorithm to fill in any holes within the segmented components. For the last step of the lung segmentation process, we used custom models to train our neural networks by applying the focal Tversky method to all four layers. In the second phase of our radiomic data analysis workflow, we attempted to differentiate patient subgroups according to specific radiomic features. Data extraction was achieved by the PyRadiomics Python package. Additionally, the Scikit-learn Python module was utilized for data processing.6 Patients were classified based on their radiomic features by applying Random Forest classification decision trees. The final decision was made by aggregating the results of all decision trees, which helps to reduce overfitting and improve the accuracy of the classification. To determine the optimal set of parameters, the Grid Search hyperparameter optimization technique was used.
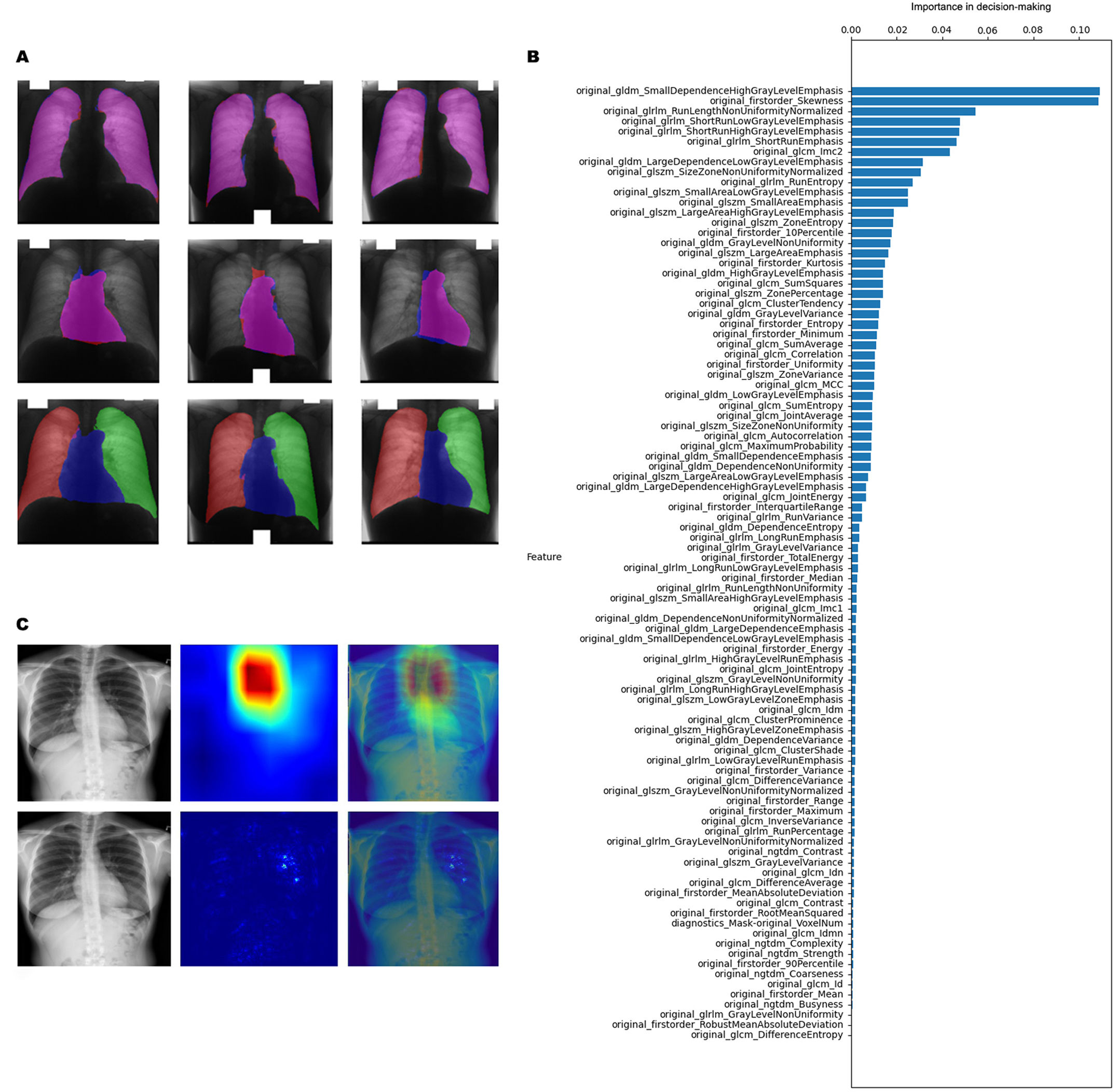
During the segmentation procedures, the algorithm accurately identified the lungs, the heart, and the background (Fig. 1A). Elevated CTR was detected in 4.84%, 6.27%, and 23.63% of all cases concerning non-COVID, post-COVID, and COVID-19 patients, respectively. Of note, the high occurrence of elevated CTR cases in the COVID-19 subgroup might be attributed to technical issues since these images were obtained in AP view instead of PA view. Although elevated CTR was detected more frequently in post-COVID patients (vs. non-COVID individuals), we found no significant differences (p=0.448) in CTR distribution concerning the abovementioned patient groups. In-depth radiomic analysis identified 96 radiomic features (Fig. 1B) that could distinguish post-COVID from COVID-19 patients and non-COVID individuals. Of note, after reducing the overfitting with the Grid Search technique, the models’ accuracy was 67.83% on the training set and 69.59% on the test set. Next, we trained another Random Forest model using a smaller subset of the most essential attributes to reduce runtime and overfitting. Parameters for this smaller dataset were selected based on their significance in decision-making, specifically targeting those with an importance score exceeding 1%. Accordingly, we downscaled the number of critical parameters for accurate classification to 28 from the original 96. The accuracy of this reduced dataset was 67.25% in the training set and 68.42% in the test set. Lastly, we aimed to visualize the regions and fields on CXRs that contributed the most to the decision-making by using the Saliency Map and GradCAM++ visualization techniques. Notably, the Saliency Map calculates the extent of pixel involvement in the decision-making process by performing backpropagation on the input image and model output. GradCAM++ is a gradient-based approach that calculates a heatmap indicating more relevant and less important areas based on a loss function. As shown in Fig. 1C, characteristic regions were identified by the algorithm that were specific for post-COVID patients. None of these areas could be detected by the human eye, further supporting the role of radiomics in comprehensive image analysis.
CXR segmentation and post-COVID-specific radiomic features. (A) Machine learning-based algorithm accurately identifies the lungs, the heart, and the background and calculates the CTR. (B) Radiomic features distinguishing post-COVID patients from COVID-19 and non-COVID individuals. (C) Visualization of representative post-COVID-specific areas on CXRs. CTR: cardiothoracic ratio; CXR: chest X-ray.
In this study, we aimed to develop machine-learning algorithms to detect post-COVID-specific radiological alterations in CXRs otherwise undetectable to the radiologists’ eyes. Although increased CTR might be linked with both respiratory and cardiac disorders,7 CTR was not diagnostic for post-COVID syndrome according to our dataset. However, when performing comprehensive radiomic profiling, we identified a panel of CXR-based radiomic features that might aid in differentiating post-COVID patients from non-COVID individuals and from acute COVID-19 patients. Upon visualization of the areas corresponding to these features, we found that the most characteristic regions were the paratracheal and the peripheral areas of the lungs. Although no additional analyses could be performed within the framework of the current study to reveal the morphological and pathophysiological aspects distinguishing these areas, we speculate that their localization might be suggestive of persistent systemic inflammation and peripheral air trapping.8 Indeed, SARS-CoV-2 infection might be associated with subepithelial inflammatory lymphomonocyte infiltrates and even vasculitis of small subepithelial vessels associated with foci of coagulative necrosis in the trachea that might persist over time.9 The diagnostic algorithm's accuracy of 69.59% is considered moderate for a clinical screening study. Nevertheless, given that CXR-based radiomics is still in its early stages of development, the results are encouraging, as they demonstrate the potential of radiomics to differentiate post-COVID patients from other groups. Incorporating larger datasets, optimizing feature selection, and refining the machine-learning algorithms will likely improve model performance and accuracy for future applications.
Although a total of 973 individuals were enrolled, this number is still considered moderate for machine learning-based studies. Accordingly, the main study limitation is the relatively low number of patients included. Chronic lung disease often mimics the symptomatology of post-COVID patients; therefore, differentiating these two entities is of clinical importance. Nevertheless, the lack of an auxiliary study group consisting of healthy (i.e. without any forms of lung disease) individuals definitely constitute a further limitation and foreshadow the need for complementary studies. Of note, including these individuals in the current study was not feasible due to radioprotective and ethical considerations, as radiography should only be performed when clinically indicated, as per institutional guidelines. When assessing specific radiomic features, the algorithm was trained to mitigate bias associated with imaging types. However, using AP imaging to determine the CTR in acute COVID-19 patients constitutes an additional limitation.
In summary, the current study is the first to distinguish specific CXR-based radiomic features diagnostic for post-COVID syndrome. Further validation of these radiomic markers might aid in the detection of these somewhat enigmatic post-acute COVID sequelae.
CRediT Authorship Contribution Statement(I) Study conception and design: Anita Rozsas, Gyorgy Marko-Varga, Balazs Dome, Anna Kerpel-Fronius, Zsolt Megyesfalvi; (II) Administrative support: All authors; (III) Provision of study materials: Anita Rozsas, Gyorgy Marko-Varga, Balazs Dome, Anna Kerpel-Fronius, Zsolt Megyesfalvi; (IV) Collection and assembly of data: Andrea Fulop, Anita Rozsas, Diana Solymosi, Bence Ferencz, Zsolt Megyesfalvi; (V) Data analysis and interpretation: Andrea Fulop, Diana Solymosi, Anna Kerpel-Fronius, Zsolt Megyesfalvi; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors.
Declaration of Generative AI and AI-assisted Technologies in the Writing ProcessNot applicable.
FundingThis research was funded by the Hungarian National Research, Development, and Innovation Office (TKP2021-EGA-33) and by the Hungarian Academy of Sciences (PC2022-II-19/1/2022). BD, ZM and AR were supported by additional funding from the Hungarian National Research, Development, and Innovation Office (KH130356 to BD; 2020-1.1.6-JÖVŐ to BD and ZM, and FK-143751 to BD, ZM and AR). BD was also supported by the Austrian Science Fund (FWF I3522, FWF I3977, and I4677). ZM was supported by the New National Excellence Program of the Ministry for Innovation and Technology of Hungary (UNKP-20-3, UNKP-21-3 and UNKP-23-5), and by the Bolyai Research Scholarship of the Hungarian Academy of Sciences. ZM is also the recipient of an International Association for the Study of Lung Cancer/International Lung Cancer Foundation Young Investigator Grant (2022). BF is a recipient of the Semmelweis 250+ Excellence PhD Scholarship (EFOP-3.6.3-VEKOP-16-2017-00009) of the Semmelweis University.
Competing InterestsThe authors declare no potential conflicts of interest.
The authors thank SuperFronix Laboratories Kft. (Budapest, Hungary) for their contribution to the radiomic analysis. We are also grateful to all the patients involved in the study and their families.