Explorations have been progressing in decoding the mechanism of non-small cell lung cancer (NSCLC). However, long noncoding RNA small nucleolar RNA host gene 5/microRNA-181c-5p/chromobox protein 4 (SNHG5/miR-181c-5p/CBX4) axis-oriented mechanisms in NSCLC is still in infancy. Therein, this study is proposed to probe this axis in NSCLC progression.
MethodsSamples of 86 NSCLC patients were collected and SNHG5, miR-181c-5p and CBX4 expression was detected in NSCLC tissues and cells. NSCLC cells were transfected with plasmids to change SNHG5, miR-181c-5p or CBX4 expression, after which cell functions and phosphorylated (p)-nuclear factor (NF)-κB protein expression were evaluated. The relationships among SNHG5, miR-181c-5p and CBX4 were validated. Tumor xenografts were implemented to verify the roles of SNHG5, miR-181c-5p and CBX4 in tumor growth.
ResultsLow miR-181c-5p and high SNHG5 and CBX4 levels were found in NSCLC tissues and cells. Restoration of miR-181c-5p or knockdown of SNHG5 or CBX4 restrained NSCLC cell progression and inactivated the NF-κB pathway. Upregulated CBX4 abolished the effects of miR-181c-5p on reducing NSCLC cell progression. SNHG5 regulated the interaction between miR-181c-5p and CBX4. In vivo, restoration of miR-181c-5p or knockdown of SNHG5 or CBX4 retarded the tumor growth.
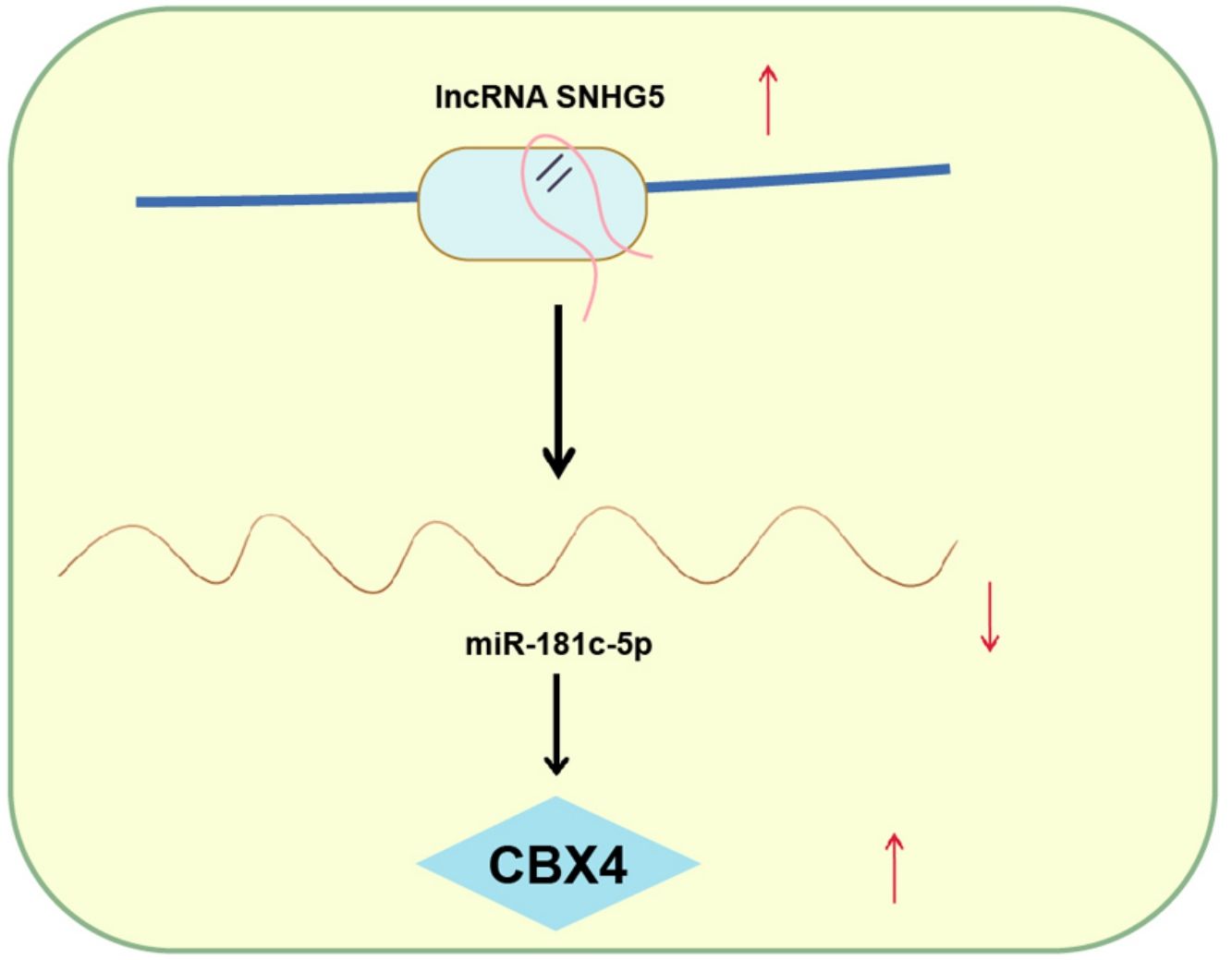
ConclusionThis study has delineated that SNHG5 induces the NF-κB pathway by regulating the miR-181c-5p/CBX4 axis to promote NSCLC progression, which may pave a novel path for NSCLC treatment.
Over the past several decades, lung cancer occupies a higher mortality rates of cancers,1 among which non-small cell lung cancer (NSCLC), accounting for more than 80% of all lung cancer cases, is categorized into adenocarcinoma, squamous cell carcinoma and large cell carcinoma.2 Regionally, the major resource of NSCLC in China is ascribed to smoking and air pollution.3 Regardless of the advancements in lung cancer treatment, chemotherapy remains the first-line option for advanced lung cancer.4 Unexpectedly, the prognosis of NSCLC is not satisfactory due to resistance to chemotherapy, resulting in recurrence of aggressive lung cancer.5 Thus, the need to discover novel therapy for NSCLC patients is in urgency.
Amount of long noncoding RNAs (lncRNAs) are differentially expressed in lung cancer.6 For instance, differentiation antagonizing noncoding RNA,7 TUC3388 and metastasis-associated lung adenocarcinoma transcript 19 are abnormally overexpressed in NSCLC, augmenting the development of the disease. LncRNA small nucleolar RNA host gene 5 (SNHG5) has been found to be a new budding star in human cancers, and be a promising diagnostic marker for cancer patients.10 In fact, SNHG5 has been surveyed to be upregulated in lung adenocarcinoma (LUAD) and be related to gefitinib resistance,11 but its related mechanism still deserves deep investigation in NSCLC. Supportive evidence has disclosed the interactions between lncRNAs and microRNAs (miRNAs) participating in lung tumorigenesis.12 In our research, the database forecasted the targeting sites between SNHG5 and miR-181c-5p. miR-18113 and miR-181b14 have been disclosed to mediate drug resistance in lung cancer. As to miR-181c-5p, it has been developed to a prognostic signature in LUAD,15 but its accurate role in NSCLC was rarely probed. miRNAs bind to 3′untranslated region (UTR) of their target mRNA to inhibit expression at the post-transcriptional level.16 Also, the binding sites of miR-181c-5p and chromobox protein 4 (CBX4) were also forecasted by online database. CBX4 is a member of the polycomb group family of epigenetic regulatory factors that has become a potent target in the control of cancer activities.17,18 Indeed, CBX4 has been verified to involve in lung cancer metastasis,19 but its regulation by miR-181c-5p in cancer progression was little mentioned. Nuclear factor (NF)-κB pathway is a cell survival pathway fanning carcinogenesis in lung cancer.20,21 NF-κB pathway is generally activated in lung cancer, and the obstruction of this pathway is critically significant to slow down lung cancer growth.22,23 Therein, this study is implemented to unearth the mechanisms of SNHG5/miR-181c-5p/CBX4 axis through the NF-κB pathway in NSCLC.
Materials and methodsEthics statementThis study was approved by the ethics committee of Sun Yat-sen University Cancer Center (ethical number: 20190608) and informed consent was acquired from patients. All animal experiments, ratified by the animal ethics committee of Sun Yat-sen University Cancer Center (ethical number: 20190814), complied with the rules and regulations of experimental animals and the relevant ethical requirements of experimental animals.
SpecimensSamples of 86 NSCLC patients were obtained from Sun Yat-sen University Cancer Center. Of these patients, 51 cases were males and 35 cases were females, aged 48–69 (58.23±3.44) years. There were 38 cases aged≤60 years, and 48 cases aged>60 years. Inclusion criteria: NSCLC diagnostic criteria. The staging system for NSCLC was the seventh edition of the TNM staging edited by International Association for the Study of Lung Cancer: 30 patients in stage I, 25 in stage II, 22 in stage III, and 9 in stage IV. Pathological type: 38 cases of adenocarcinoma, 31 cases of squamous cell carcinoma, and 17 cases of large cell carcinoma. There were 49 cases of lymph node metastasis. All of the patients were clinically diagnosed and did not receive chemoradiotherapy before surgery. The surgically excised cancer and normal adjacent tissues (>5cm from the edge of cancer tissues) were adopted.
Cell cultureHuman embryo lung cells (MRC-5), and human NSCLC cell lines (HCC827, H1299, H1975 and A549) were provided by Cell Bank of Chinese Academy of Sciences (Shanghai, China). These cells were cultured in corresponding complete mediums containing 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin (Corning Incorporated, Corning, NY, USA). Experimental cells were all in the logarithmic growth phase. Roswell Park Memorial Institute 1640 (RPMI-1640) cell culture medium, minimum essential medium (MEM), and Ham's F-12K medium were provided by Thermo Fisher Scientific (Waltham, MA, USA).
Cell transfection and groupingSmall interfering RNA (siRNA) against SNHG5 (si-SNHG5, final concentration of 20nM) and its matched negative control (si-NC, final concentration of 20nM), miR-181c-5p mimic (final concentration of 10nM) and its matched NC (mimic NC, final concentration of 10nM), siRNA targeting CBX4 (si-CBX4, final concentration of 20nM) and its matched NC (si-con, final concentration of 20nM) were synthesized in GenePharma (Shanghai, China). The fragments of CBX4 were inserted into pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA) for the construction of overexpression plasmid (oe-CBX4, final concentration of 1μg/mL). A549 cells were transfected with the aforesaid various constructs as appropriate using the Lipofectamine™ 3000 Transfection Reagent. The medium was replaced 6h post transfection, and the cells were obtained 48h later.
Cell counting kit (CCK)-8 assayThe A549 cells (4×103cells) were seeded in 96-well plates and adhered to the walls. After 24, 48, and 72h of incubation, 10μL CCK-8 reagent was added to each well and the absorbance (A) values at 490nm were read after 2h of CCK-8 treatment. Three replicates were set for each treatment and time point.24
Colony formation assayA549 cells were detached with concentration adjustment. The cell solution (10mL, 300cells) was seeded into a 6-cm petri dish and incubated for 10d. Then, the cells were deprived of culture medium and fixed by paraformaldehyde. Followed by that, the cells were stained by crystal violet solution, rinsed with tap water and photographed to calculate the number of colonies in each culture dish.
Flow cytometryTransfected A549 cells were resuspended in 1× Annexin binding buffer (500μL) to acquire a concentration of 1×106cells/mL, followed by dying with 5μL of Annexin V-fluorescein isothiocyanate (FITC) (BD Biosciences) and 10μL of propidium iodide (PI). The Annexin-V-PI double negative group (unstained cells), Annexin-V-single staining group (Annexin-V-FITC stained cells alone) and PI single stained group (PI stained cells alone) were set as references. The flow cytometer was set with the excitation wavelength at 488nm, and the emission wavelength at 530nm FL1 was the green fluorescent of FITC channel while FL2 was red fluorescence of PI channel.
With Annexin-V as the horizontal axis and PI as the vertical axis, the upper left quadrant (Annexin-V-, PI+) stood for necrotic cells, the lower left quadrant (Annexin-V-, PI−) for living cells, the upper right quadrant (Annexin-V+, PI−) for early apoptotic cells and the lower right quadrant (Annexin-V+, PI+) for late apoptotic cells.25
Transwell assayResuspended in serum-free culture medium to an optimal concentration, A549 cells (5×104cells) were seeded in a Matrigel-coated Transwell chamber at 500μL/well in the bottom of the chamber and supplemented with 10% FBS for 30-h of incubation. After that, the cells on the chamber were wiped with cotton swabs, stained by crystal violet staining solution, as well as captured in each field under a microscope. The chamber in the migration experiment was not coated with Matrigel, and the remaining steps were the same as the invasion experiment.26
Tumor xenografts in nude miceThe Lentiviral Packaging kit (Thermo Fisher Scientific) was utilized for stably knocking down SNHG5, overexpressing miR-181c-5p, as well as knocking down CBX4 in A549 cells. Lentivirus carrying si-SNHG5, miR-181c-5p mimic, si-CBX4 and their matched NCs were all packaged following the manufacturer's requirements. A549 cells were transfected with lentivirus carrying si-SNHG5, miR-181c-5p mimic, si-CBX4 and their matched NCs in the presence of polybrene (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and selected by puromycin (Sigma-Aldrich; Merck KGaA) for 2w to obtain the stable cell lines.
Forty-eight BALB/c nude mice (6–8 weeks old) were raised in a sterilized animal room of specific pathogen-free grade (25°C, 60–70% humidity) and were free to food and water. The nude mice were randomly classified into several groups (n=8/group): si-NC, si-SNHG5, mimic NC, miR-181c-5p mimic, si-con and si-CBX4 groups, Based on the groupings, the stably-transfected A549 cell suspension (2×106) was injected subcutaneously into the right back of nude mice. Each nude mouse was marked with a marker pen. When a tumor was palpable, the longest diameter (length) and shortest diameter (width) were measured with an ultraviolet-sterilized ruler and the measurement was performed every 7d. Tumor volume was calculated as π/6×(length×width).2 After measuring the tumor size for 4w, the nude mice were euthanized by CO2 euthanasia and their subcutaneous tumors were removed, photographed and weighed.27
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)Trizol kit (Invitrogen, CA, USA) was in application for RNA extraction from tissues and cells. RNA was dissolved in RNase-free water, and detected by a DU-800 accounting protein analyzer (Beckman Coulter Life Sciences, Brea, CA, USA) for RNA purity and concentration detection. For the quantification of SNHG5 and CBX4, reverse transcription was performed using HiScript® II reverse transcriptase (Vazyme, Nanjing, China) and GAPDH was used as the internal reference. For miR-181c-5p, miRNA First-Stand cDNA Synthesis Kit (Vazyme) was used, with U6 as the internal reference. PCR primers are listed in Supplementary Table 1. The data was processed by the 2−ΔΔCt method.
Western blot analysisTotal tissue and cell proteins were extracted by a radio-immunoprecipitation assay kit (Beyotime, Shanghai, China). Protein concentration was measured by a bicinchoninic acid protein concentration assay kit. The protein sample was loaded, separated by 10% SDS-PAGE and electroblotted onto a membrane. Followed by that, the protein membrane was treated with 5% skim milk powder, probed with CBX4 (1:1000), phospho (p)-NF-κB p65 (Ser536) (1:1000), GAPDH (1:1000; all from CST, MA, USA) and reprobed with corresponding secondary antibody. The protein bands were evaluated for the gray values by gel graphic analysis software Image Lab.
Dual luciferase reporter gene assayThe sequences of SNHG5 or 3′UTR of CBX4 including the wild-type or mutant binding sites of miR-181c-5p were amplified and cloned into pmirGLO plasmid (Promega, Fitchburg, WI, USA) to generate the luciferase plasmids. miR-181c-5p mimic and its NC were transfected into A549 cells together with constructed vectors (SNHG5-WT, SNHG5-MUT, CBX4-3′UTR WT or CBX4-3′UTR MUT). Cells were lysed and the luciferase activity was detected by a luciferase reporter system (Promega, WI, USA).28
Statistical analysisSPSS 21.0 (IBM Corp. Armonk, NY, USA) and GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA) were utilized for data evaluation. Measurement data were presented as mean±standard deviation. Comparisons between two groups were assessed by t-test while those among multiple groups by one-way analysis of variance (ANOVA) and Tukey's poct hoc test. Pearson correlation analysis was adopted. Enumeration data were expressed as rates or percentages, and Fisher's exact test was indicated to comparative analysis. p<0.05 was considered of statistical significance.
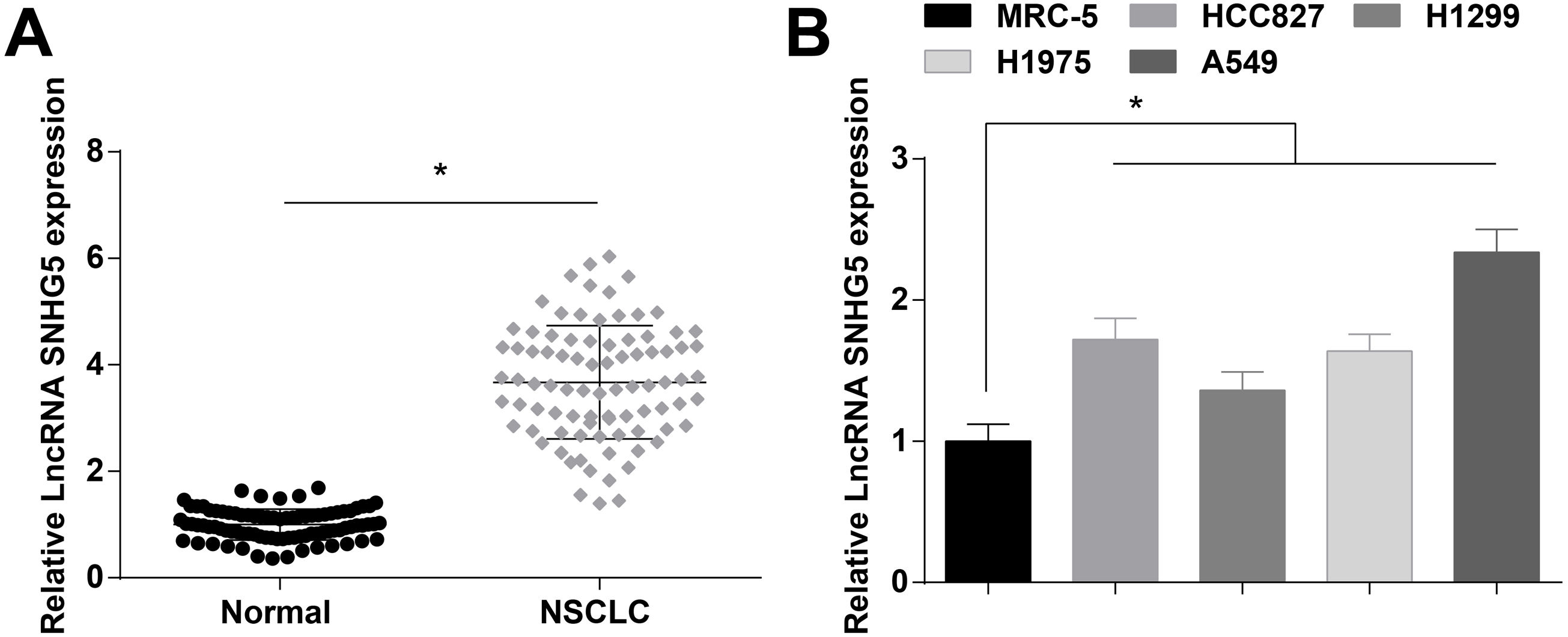
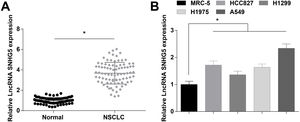
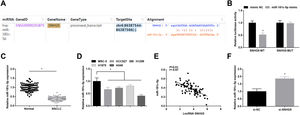
ResultsSNHG5 is highly expressed in NSCLCSNHG5 levels in clinical tissue samples and cell lines was detected by RT-qPCR, and it was found that SNHG5 was upregulated in NSCLC tissues (Fig. 1A) and in cells lines (HCC827, H1299, H1975 and A549) (Fig. 1B). According to the histopathological classification, NSCLC could be classified into adenocarcinoma, squamous cell carcinoma and large cell carcinoma, while the expression levels of SNHG5, miR-181c-5p and CBX4 were not significantly different in adenocarcinoma, squamous cell carcinoma and large cell carcinoma patients (Supplementary Fig. 1). SNHG5 upregulation was especially obvious in A549 cells, thus follow experiments were performed using A549 cells.
Patients were divided into a high and a low expression groups according to the median values of SNHG5 expression levels. Patients in the two groups were subjected to clinicopathological analysis, and the outcomes disclosed that the advanced TNM staging, lymph node metastasis, and poorly differentiated tumors were correlated to high expression of SNHG5 (all p<0.05), while SNHG5 had no correlation with age, gender, and tumor diameter (all p>0.05) (Supplementary Table 2).
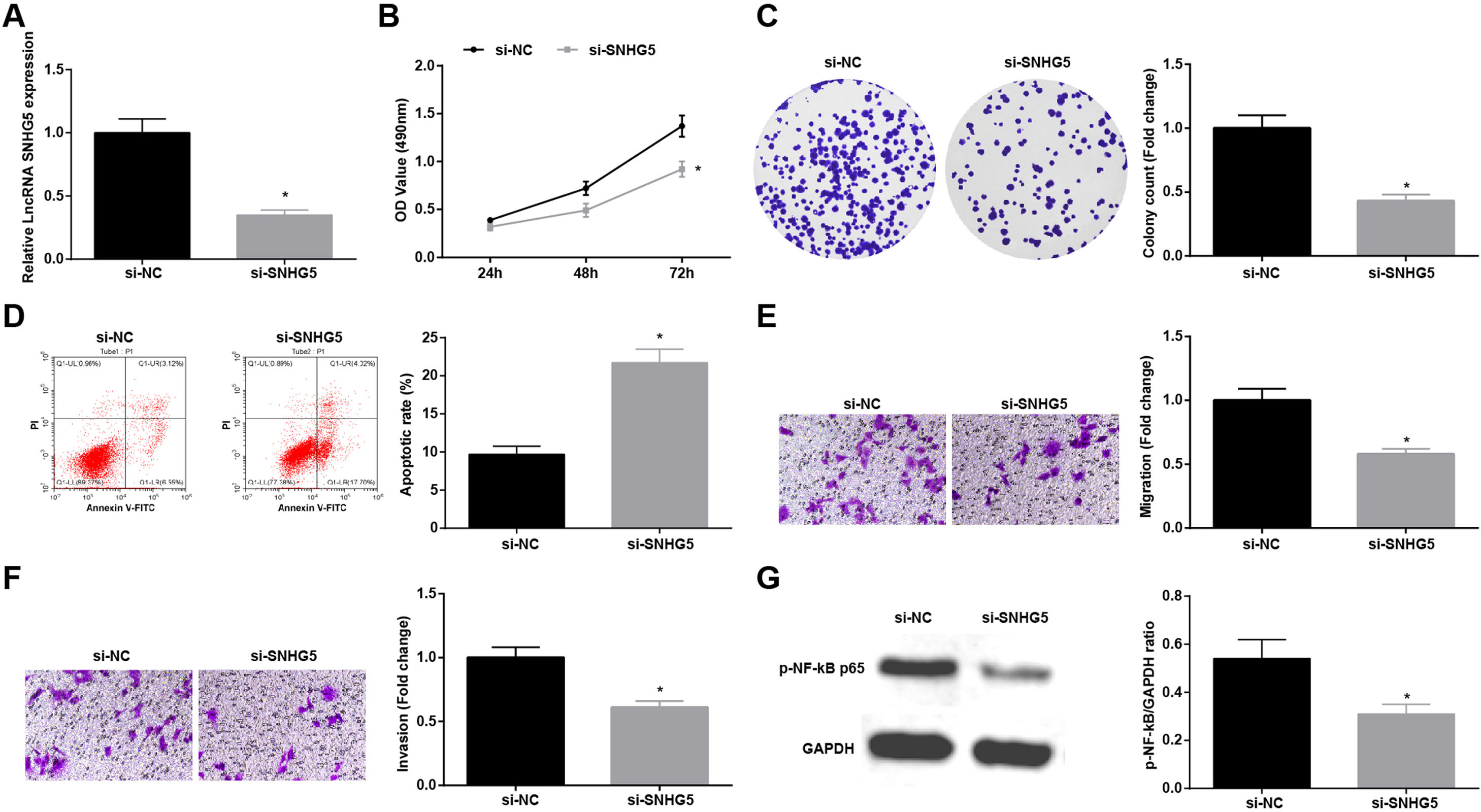
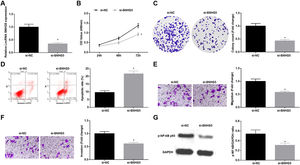
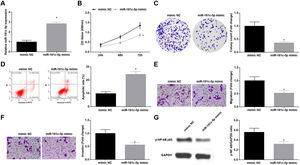
Silenced SNHG5 retards NSCLC progressionTo explore the effect of SNHG5 induced the NF-κB signaling pathway on NSCLC, si-SNHG5 was introduced into A549 cells to downregulate SNHG5 expression (Fig. 2A). CCK-8, colony formation assay, flow cytometry and Transwell assay were utilized to discover that by downregulation of SNHG5, A549 cell proliferation, colony formation, invasion and migration capacities impaired and apoptotic rate increased (Fig. 2B–F). Also, Western blot found that after SNHG5 downregulation, p-NF-κB-p65 protein levels were decreased in A549 cells (Fig. 2G).
Silenced SNHG5 reduces the development of NSCLC cells. (A) SNHG5 expression in A549 cells was detected by RT-qPCR; (B) A549 cell proliferation after SNHG5 silencing was assessed by CCK-8 assay; (C) Colony formation rate of A549 cells after SNHG5 silencing was tested by colony formation assay; (D) A549 cell apoptosis rate after SNHG5 silencing was evaluated by flow cytometry; (E) A549 cell migration after SNHG5 silencing was examined by Transwell assay; (F) A549 cell invasion after SNHG5 silencing was determined by Transwell assay; (G) p-NF-κB p65 protein expression in A549 cells after SNHG5 silencing was measured by Western blot. Measurement data were expressed as mean±standard. * P<0.05 vs. the si-NC group. Cell experiments were performed independently in triplicate.
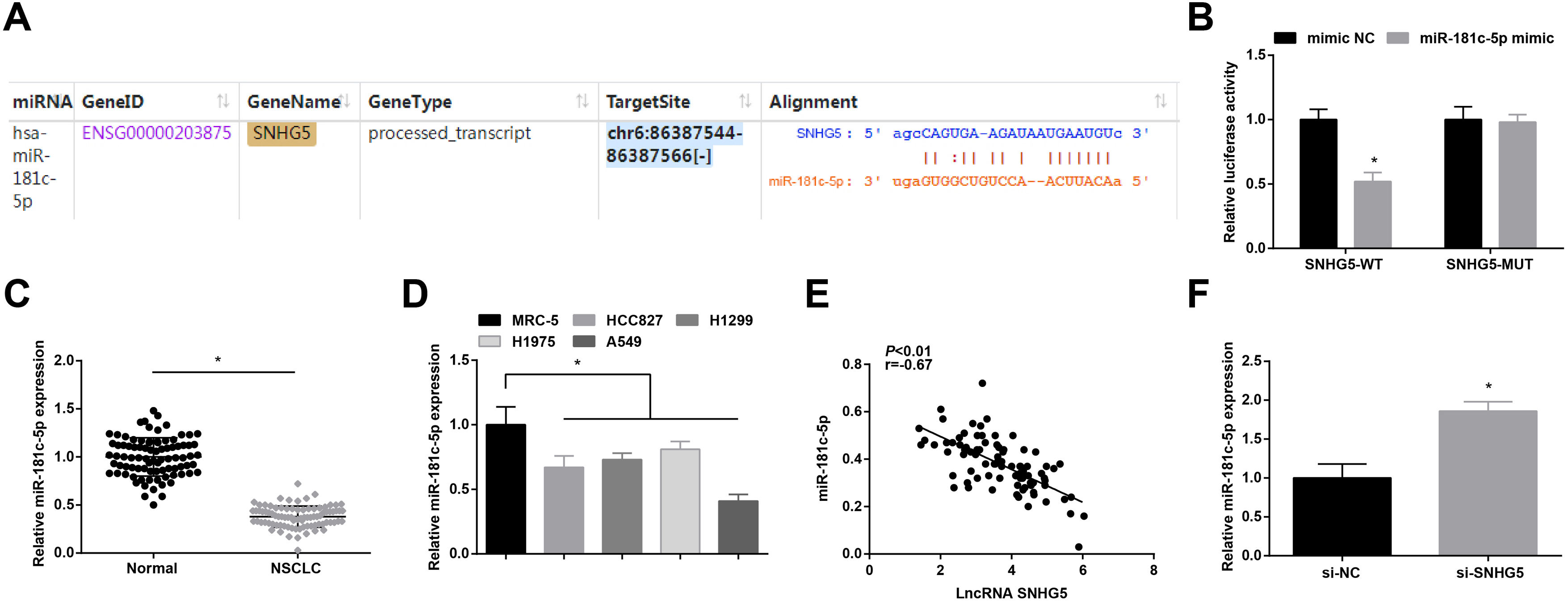
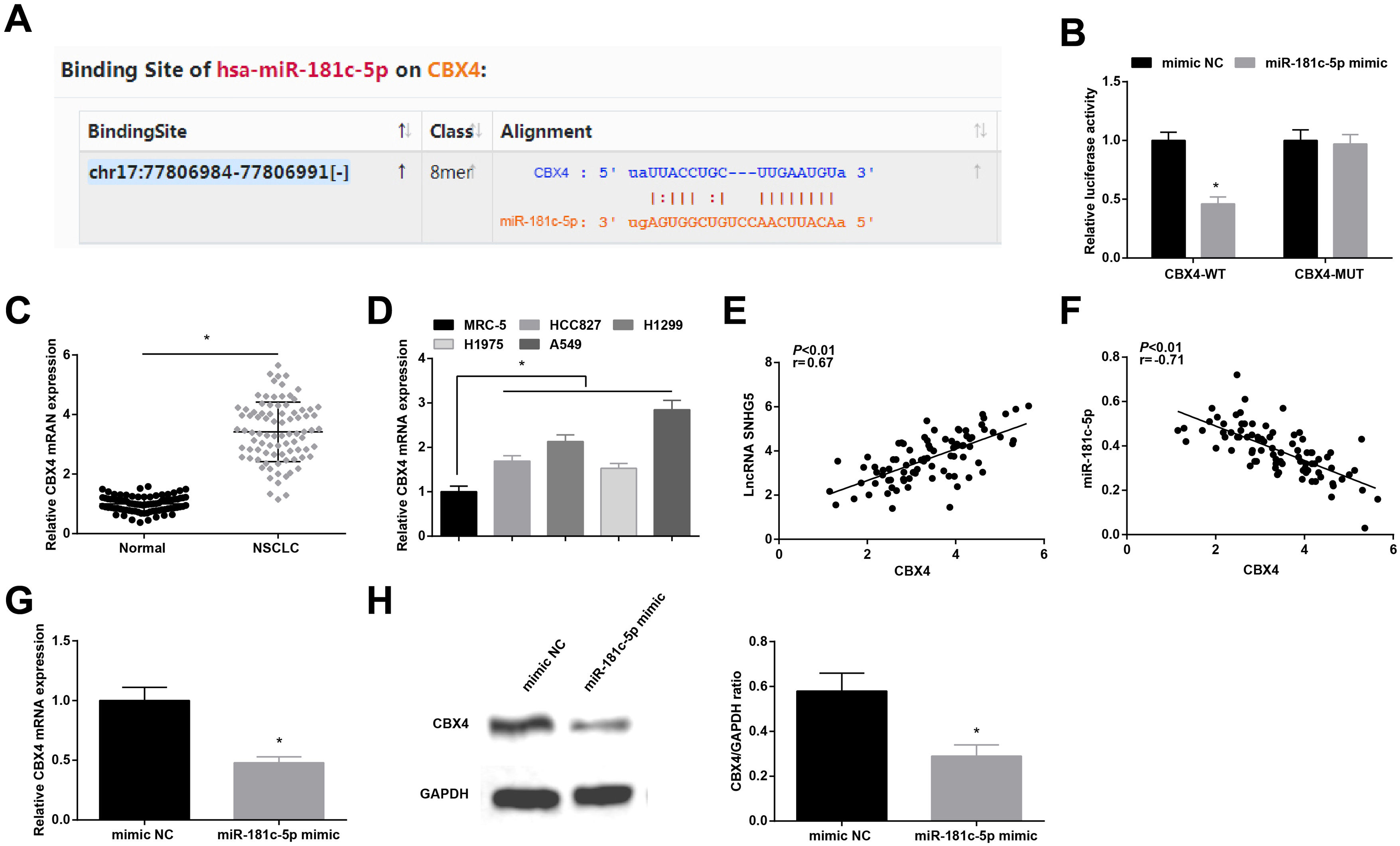
Starbase (http://starbase.sysu.edu.cn/index.php) indicated the targeting sites between SNHG5 and miR-181c-5p (Fig. 3A). Dual luciferase reporter gene assay examined that A549 cells co-transfected with SNHG5-WT and miR-181c-5p mimic had impaired luciferase activity (Fig. 3B), revealing that SNHG5 could directly bind to miR-181c-5p.
SNHG5 binds to miR-181c-5p. (A) Binding sites of SNHG5 and miR-181c-5p were analyzed by Starbase; (B) The binding of SNHG5 to miR-181c-5p was assessed by luciferase activity assay; (C, D) miR-181c-5p expression in clinical tissues and cell lines was tested by RT-qPCR; (E) Pearson analysis of SNHG5 and miR-181c-5p; (F) miR-181c-5p expression in A549 cells after SNHG5 silencing was tested by RT-qPCR. Measurement data were expressed as mean±standard. In (B), * P<0.05 vs. mimic NC group (N=3); in (C), * P<0.05 vs. normal tissues (n=86); in (D), * P<0.05 vs. MRC-5 cells (N=3); in (F), * P<0.05 vs. si-NC group (N=3).
In NSCLC tissues and cells, miR-181c-5p expression was tested to be decreased (Fig. 3C, D). In clinical cancer tissues, Pearson correlation analysis showed that SNHG5 was negatively correlated with miR-181c-5p levels (Fig. 3E). Moreover, in A549 cells after silencing SNHG5, miR-181c-5p was found to be upregulated (Fig. 3F).
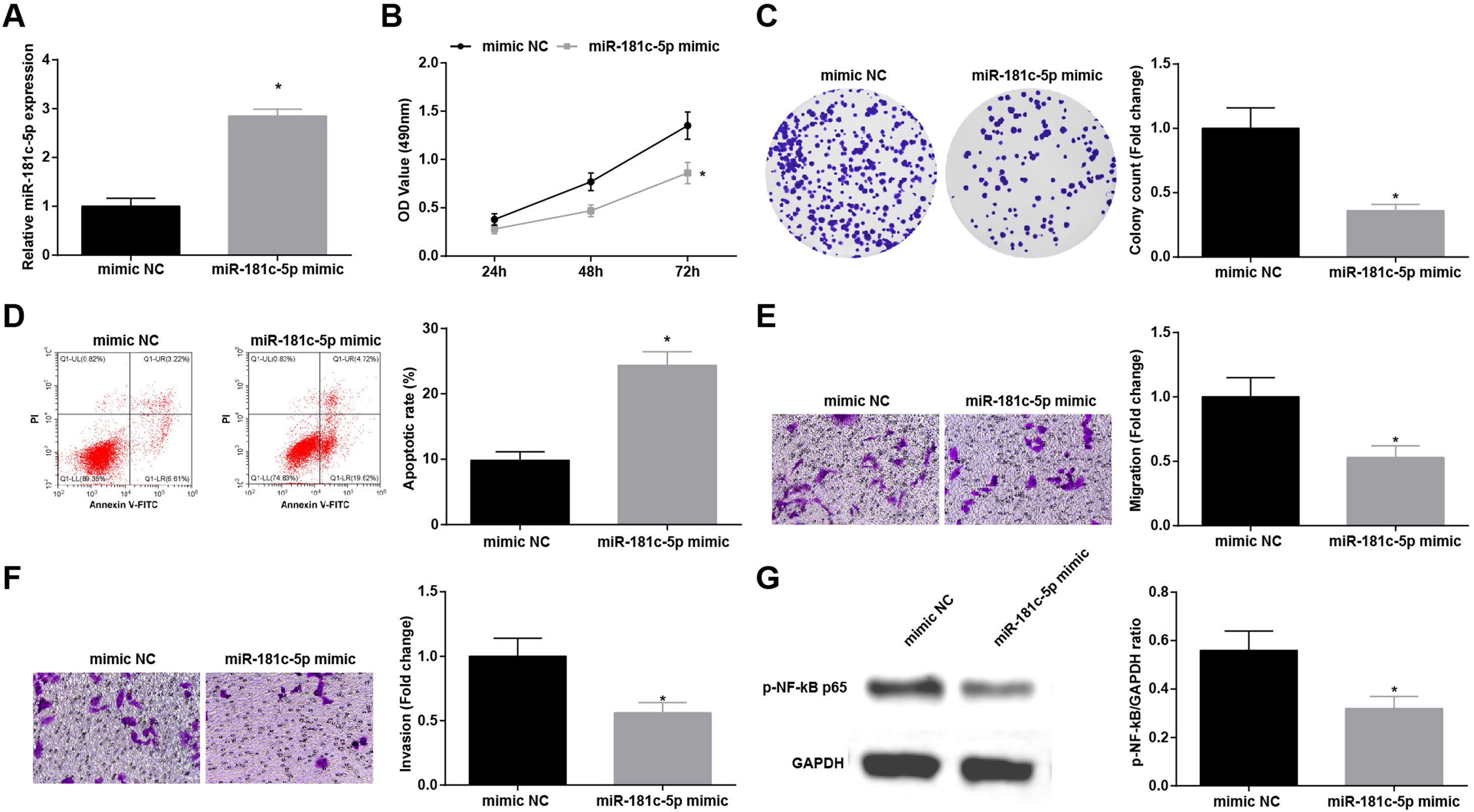
Restoration of miR-181c-5p restrains NSCLC cell progressionTo explore the influence of miR-181c-5p on NSCLC, A549 cells were treated with miR-181c-5p mimic to enrich miR-181c-5p expression (Fig. 4A). Then, in A549 cells expressing enriched miR-181c-5p, it was manifested that cell proliferation, colony formation, invasion and migration capacities impaired and apoptotic rate increased (Fig. 4B–F), and p-NF-κB-p65 protein expression decreased (Fig. 4G).
Restoration of miR-181c-5p restrains NSCLC cell progression. (A) miR-181c-5p expression in A549 cells was detected by RT-qPCR; (B) A549 cell proliferation after upregulating miR-181c-5p was assessed by CCK-8 assay; (C) Colony formation rate of A549 cells after upregulating miR-181c-5p was detected by colony formation assay; (D) A549 cell apoptosis rate after upregulating miR-181c-5p was tested by flow cytometry; (E) A549 cell migration after upregulating miR-181c-5p was detected by Transwell assay; (F) A549 cell invasion after upregulating miR-181c-5p was evaluated by Transwell assay; (G) p-NF-κB p65 protein expression in A549 cells after upregulating miR-181c-5p was measured by Western blot. Measurement data were expressed as mean±standard. * P<0.05 vs. the mimic NC group. Cell experiments were performed independently in triplicate.
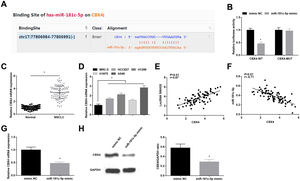
Starbase predicted the binding sites of miR-181c-5p and CBX4 (Fig. 5A). Dual-luciferase reporter experiment further detected the binding relationship between miR-181c-5p and CBX4, as the results demonstrating that miR-181c-5p mimic diminished the luciferase activity of CBX4-WT in A549 cells (Fig. 5B), suggesting that miR-181c-5p could directly target CBX4.
miR-181c-5p targets CBX4. (A) Binding sites of miR-181c-5p and CBX4 were predicted by Starbase; (B) The binding of miR-181c-5p and CBX4 was verified by luciferase activity assay; (C, D) CBX4 expression in clinical tissues and cell lines was tested by RT-qPCR; (E, F) Pearson correlation analysis of SNHG5, miR-181c-5p and CBX4; G-H. CBX4 expression was measured by RT-qPCR and western blot. Measurement data were expressed as mean±standard. In (B, G and H), * P<0.05 vs. mimic NC group (N=3); in (C), * P<0.05 vs. normal tissues (n=86); in (D), * P<0.05 vs. MRC-5 cells (N=3).
High CBX4 mRNA levels were examined in NSCLC tissues and cells lines (Fig. 5C, D). Further analysis by Pearson test disclosed that CBX4 expression was positively correlated with SNHG5 while negatively correlated with miR-181c-5p (Fig. 5E, F). CBX4 mRNA and protein levels were determined to be inhibited after miR-181c-5p overexpression in A549 cells (Fig. 5G, H).
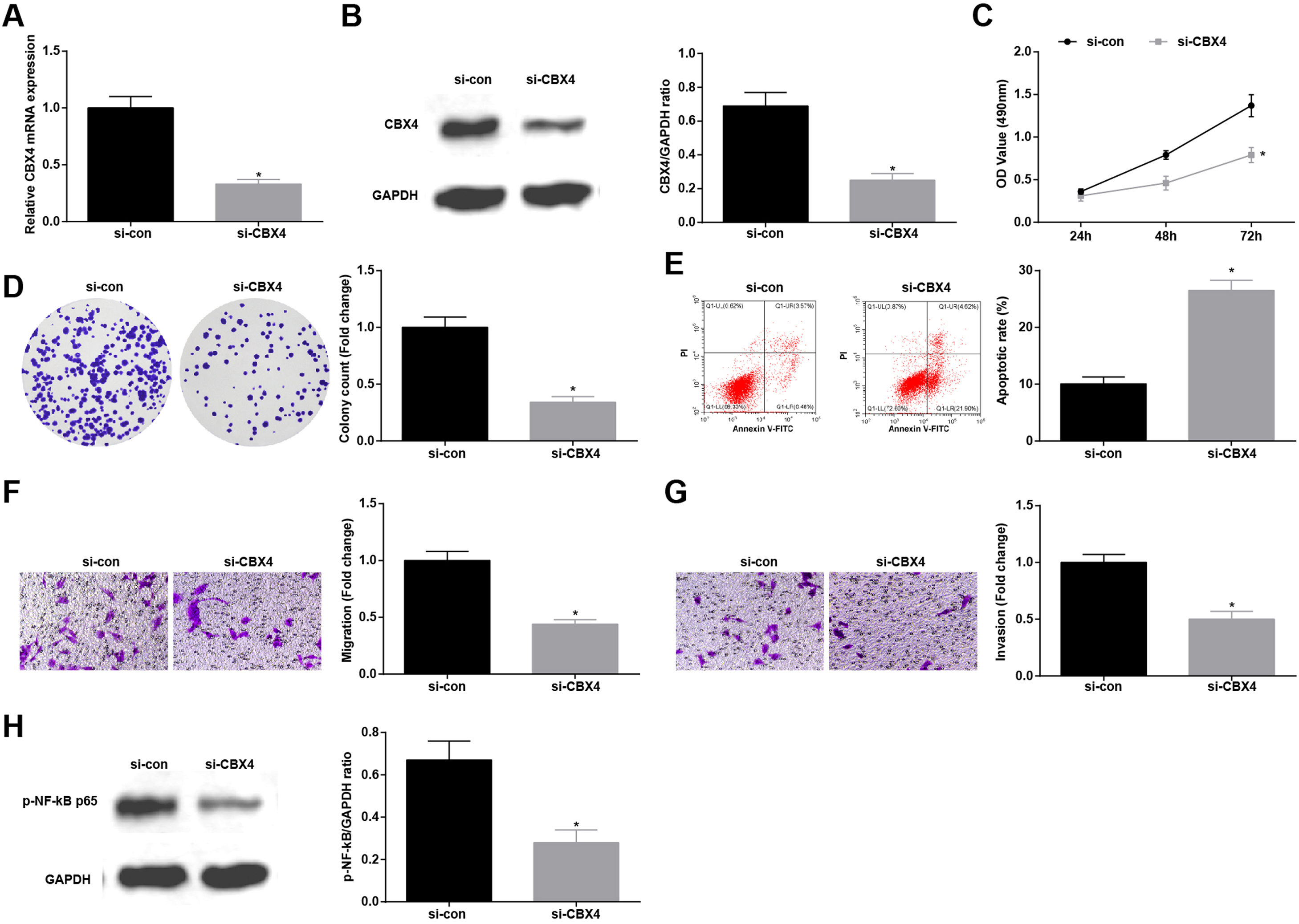
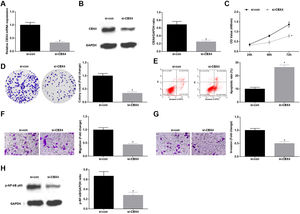
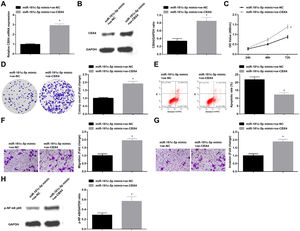
Knockdown of CBX4 suppresses NSCLC cell progressionTo prove that CBX4 also has a regulatory effect on development of A549 cells, CBX4 was knocked down in A549 cells by transfection with si-CBX4 (Fig. 6A, B). Next, it was further evaluated that CBX4 knockdown in A549 cells repressed cellular growth (Fig. 6C–G) and reduced p-NF-κB-p65 protein expression (Fig. 6H).
Knockdown of CBX4 suppresses NSCLC cell progression. (A, B) CBX4 expression in A549 cells was detected by RT-qPCR and western blot; (C) A549 cell proliferation after silencing CBX4 was detected by CCK-8 assay; (D) Colony formation rate of A549 cells after silencing CBX4 was evaluated by colony formation assay; (E) A549 cell apoptosis rate after silencing CBX4 was examined by flow cytometry; (F) A549 cell migration after silencing CBX4 was measured by Transwell assay; (G) A549 cell invasion after silencing CBX4 was detected by Transwell assay; (H) p-NF-κB p65 protein expression in A549 cells after silencing CBX4 was tested by Western blot. Measurement data were expressed as mean±standard deviation; * P<0.05 vs. the si-con group. Cell experiments were performed independently in triplicate.
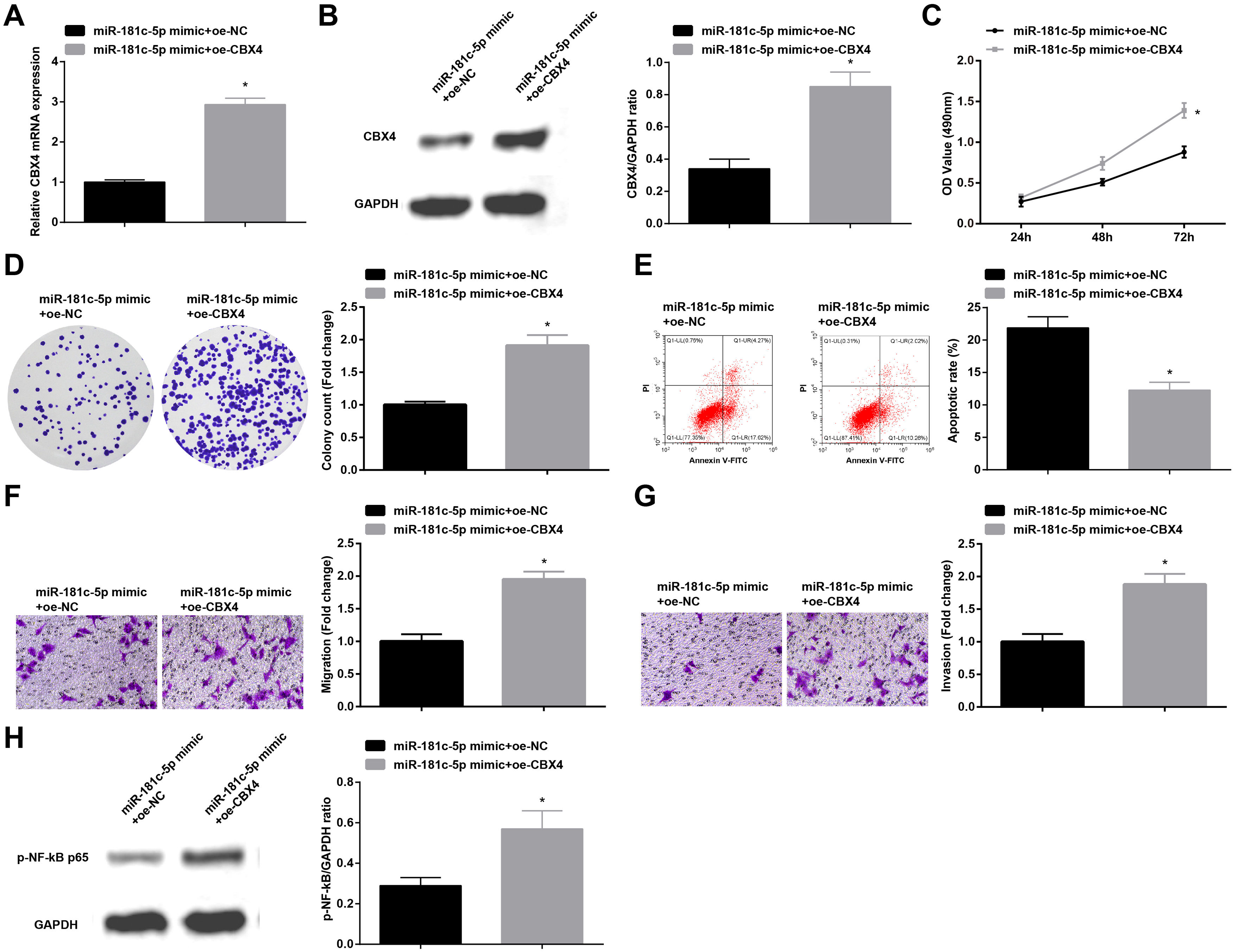
In order to identify the role of CBX4 on miR-181c-5p-induced regulation of A549 cell growth, A549 cells after transfection with miR-181c-5p mimic were further treated with Oe-CBX4. A series of experimental results unveiled that the overexpression of CBX4 reversed miR-181c-5p overexpression-induced suppression of A549 cells malignant phenotype and p-NF-B-p65 protein expression (Fig. 7A–H).
Overexpression of CBX4 abolishes the effects of miR-181c-5p on NSCLC cell progression. (A, B) CBX4 expression in A549 cells was tested by RT-qPCR and western blot; (C) A549 cell proliferation in the rescue experiment was detected by CCK-8 assay; (D) Colony formation rate of A549 cells in the rescue experiment was tested by colony formation assay; (E) A549 cell apoptosis rate in the rescue experiment was assessed by flow cytometry; (F) A549 cell migration in the rescue experiment was examined by Transwell assay; (G) A549 cell invasion in the rescue experiment was detected by Transwell assay; (H) p-NF-κB p65 protein expression in A549 cells in the rescue experiment was determined by Western blot. Measurement data were expressed as mean±standard deviation; * P<0.05 vs. the miR-181c-5p mimic+oe-NC group. Cell experiments were performed independently in triplicate.
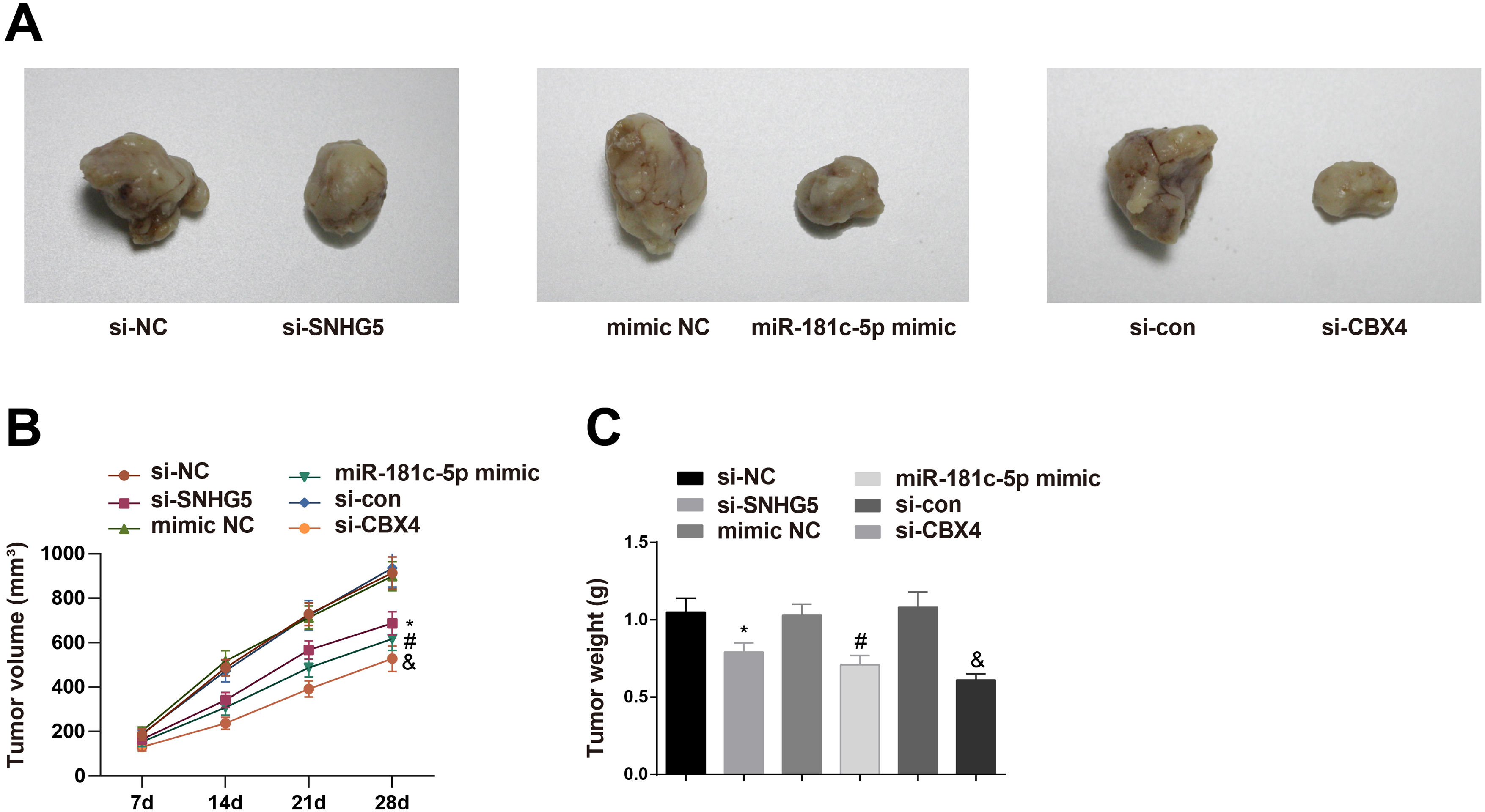
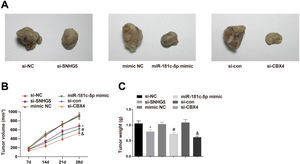
To investigate the roles of the SNHG5/miR-181c-5p/CBX4 axis in tumorigenesis in vivo, the mice xenograft model assay was conducted. The corresponding findings revealed that downregulation of SNHG5 and CBX4 or upregulation of miR-181c-5p retarded tumor growth in nude mice (Fig. 8A–C).
Effect of the SNHG5/miR-181c-5p/CBX4 axis on the development of NSCLC in vivo. (A) Representative figures for tumors. (B) Tumor growth volume after tumor transplantation of A549 cells in each group. (C) Tumor growth weight after tumor transplantation of A549 cells in each group. Measurement data were expressed as mean±standard deviation. n=8. * P<0.05 vs. the si-NC group; # P<0.05 vs. the mimic NC group; & P<0.05 vs. the si-con group.
NSCLC is still the main issue needed to be overcame based on its high mortality rate and poor prognosis.29 Through extensive researches have been projected to address the precise comprehension of the SNHG5/miR-181c-5p/CBX4 axis in NSCLC is obscure. Hence, this study is initiated and the mainstay of the outcomes holds that SNHG5 promotes NSCLC cell progression through lessening miR-181c-5p-induced regulation of CBX4. Specifically, low miR-181c-5p and high SNHG5 and CBX4 levels were found in NSCLC tissues and cells. Restoration of miR-181c-5p or knockdown of SNHG5 or CBX4 restrained NSCLC cell progression and inactivated the NF-κB pathway. Upregulated CBX4 abolished the effects of miR-181c-5p on reducing NSCLC cell progression. SNHG5 regulated the interaction between miR-181c-5p and CBX4. In vivo, restoration of miR-181c-5p or knockdown of SNHG5 or CBX4 retarded the tumor growth.
Initially, SNHG5 expression in NSCLC tissues and cell lines was determined, as well as the correlation between its expression and clinicopathological characteristics of NSCLC patients. The results indicated that high SNHG5 in NSCLC was connected with TNM stage, lymph node metastasis and high differentiation of NSCLC tissues. Increased SNHG5 has been determined in clear cell renal cell carcinoma, exhibiting an intimate correlation with TNM stage and lymph node metastasis.30 In patients with hepatocellular carcinoma, upregulated SNHG5 is found in cancer tissues, which has a correlation with patients’ TNM stage.31 Next, our research designed function assays and revealed that silenced SNHG5 hindered NSCLC cell progression and inactivated the NF-κB pathway. Accordingly, Jiang-Rui Chi et al. have observed that breast cancer cell proliferation could be induced by SNHG5 overexpression, but suppressed by SNHG5 depletion.32 Meanwhile, an article composed by Mingbao Zhang et al. has suggested that SNHG5 induction enhances proliferation, migration and anti-apoptosis of colorectal cancer cells.33 In a case of melanoma, it has been clarified that SNHG5 decrease in cancer cells results in proliferation and invasion inhibition and apoptosis promotion.34
Next, our research found the binding of SNHG5 to miR-181c-5p. Pivoted on miR-181c-5p, our study further validated that enriched miR-181c-5p exerted suppressively for NSCLC cells to behave malignantly and for the NF-κB pathway to activate. Not limited to the present paper, the decrease of miR-181c-5p has been measured in various cancers, including cervical squamous cell carcinoma.26 In the area of miR-181c-5p-mediated growth of cancer cells, Han B et al. have summarized that miR-181c upregulation hinders proliferation of drug-resistant breast cancer cells.35 On the other hand, miR-181c has acts as a tumor suppressor because of its inhibitory influences on glioblastoma cell invasion, proliferation and self-renewal properties.36 Furthermore, Yong Li et al. have noticed that neuroblastoma cells exhibit decelerated proliferation, migration and invasion when miR-181c is overexpressed.37 Paying attention to the regulation of miR-181c-5p for the NF-κB pathway, it has been once declared that miR-181 overexpression in ovarian cancer cells lowers the NF-κB expression.38
Subsequently, CBX4 that was targeted by miR-181c-5p was analyzed to be upregulated in NSCLC. Through cell function measurements, the outcomes depicted the anti-tumor function of silenced CBX4. Moreover, CBX4 overexpression counteracted miR-181c-5p overexpression-dependent suppression of NSCLC cell growth. Based on a previous article, it is known that CBX4 is highly expressed in lung cancer, and the treatment with overexpressed CBX4 accounts for augmented proliferation and migration capabilities.19 For breast cancer, CBX4 expression is discovered to be elevated, and CBX4-mediated proliferation promotion could be repressed by silencing CBX4.18 Besides, in a publication addressed by Jianfa Li et al. it is noticeable that silenced CBX4 is feasible to assist circRNA TLK1-mediated effects on suppressing phenotypic transformation of renal cell carcinoma cells.39
In generally, this study has elucidated that SNHG5 downregulation elevates miR-181c-5p expression to silence CBX4, thereby impeding NSCLC cell progression. The present analysis provides a novel reference for exploring NSCLC therapeutic agents. Except for the NF-κB pathway, other pathways may be involved in the axis of the SNHG5/miR-181c-5p/CBX4 promoting NSCLC progression, which is worthy exploring in the future.
Conflict of interestThe authors declare no competing interests.