Journal Information
Vol. 61. Issue 4.
Pages 242-244 (April 2025)
Share
Download PDF
More article options
Vol. 61. Issue 4.
Pages 242-244 (April 2025)
Discussion Letter
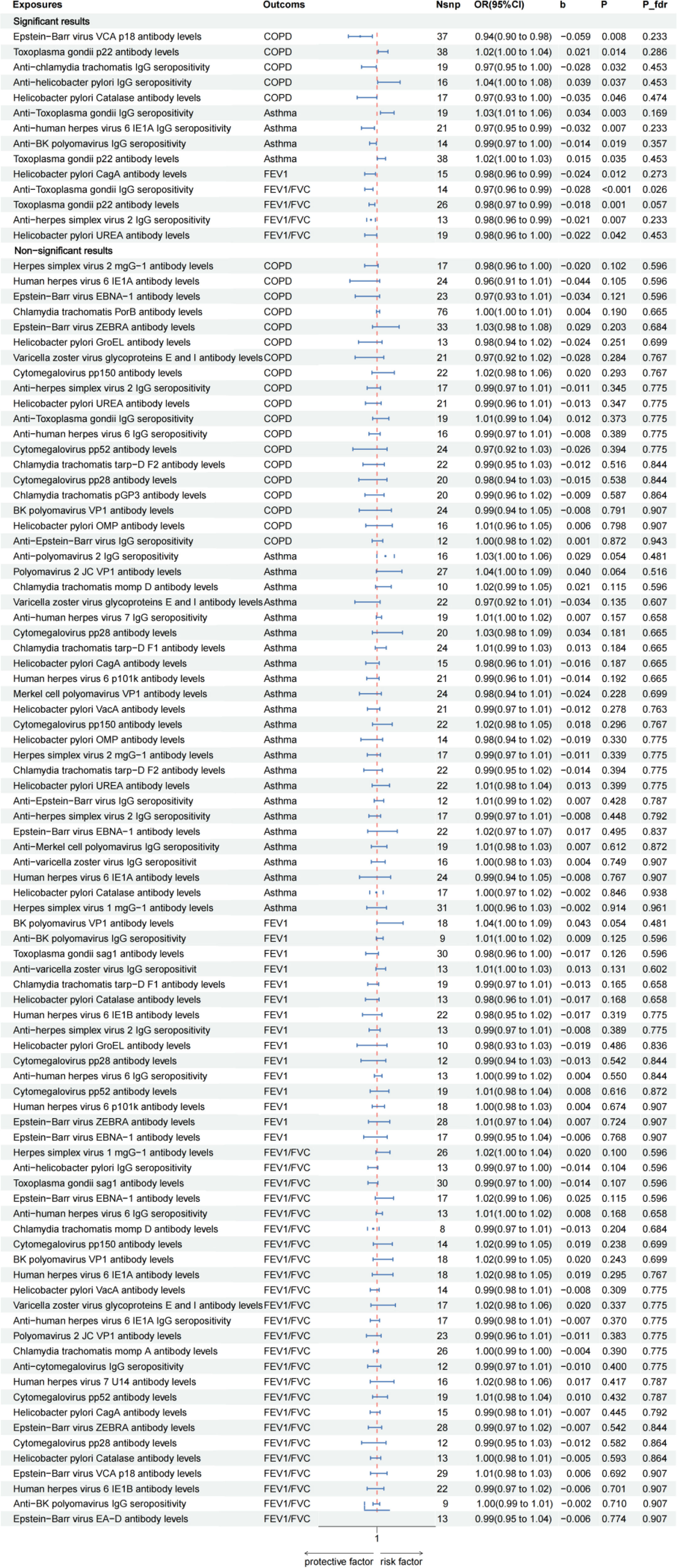
A Commentary on ‘Effect of the Antibody-Mediated Immune Responses on COPD, Asthma, and Lung Function: A Mendelian Randomization Study’
Visits
488
a Division of Endocrinology, Department of Internal Medicine, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
b Branch of National Clinical Research Center for Metabolic Disease, Wuhan, China
c Division of Cardiology, Department of Internal Medicine, Liyuan Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
This item has received
Article information
These are the options to access the full texts of the publication Archivos de Bronconeumología