Chronic obstructive pulmonary disease (COPD) is a complex syndrome with an inflammatory component, that emerges from the interaction between the genetic background of the individual and several environmental factors. Cigarette smoking (CS) is the main environmental risk factor for COPD.1,2 Yet, exposure to smoke coming from biomass burning (biomass smoke, BS) is also recognized as an important environmental COPD risk factor.3 CS and BS-COPD patients show clinical, functional, imaging, and histopathological differences suggesting that they correspond to two different clinical COPD phenotypes.4
Micro-RNAs (miRNAs) are a type of small noncoding RNAs that regulate a wide range of cellular activities. Here, we compared the levels of 2609 circulating miRNAs in patients with COPD associated to cigarette smoking and in never-smoking BS-COPD patients, exploring the hub genes and pathways associated with these differentially expressed miRNAs. 15 never-smoking COPD patients with a history of BS exposure (BS-COPD group) and 15 age and sex-matched ever-smokers with COPD (CS-COPD group, 10 ex-smokers, 5 current smokers) from a rural population in Chile were recruited at the Respiratory Service of the Regional Hospital de Talca, where they attended to undergo diagnostic tests after suspected COPD or for COPD monitoring visits. The diagnosis of COPD followed the GOLD criteria.5 Medical history was collected using standardized questionnaires including a standardized version of the CanCOLD study questionnaire,6 adding some questions referring to biomass fuels. Cumulative exposure to BS was calculated as hour-years as previously described7 and cigarette smoking history was measured by pack-years. This study was conducted ethically in accordance with the World Medical Association Declaration of Helsinki, and the Ethics Committees of the Maulean Health Service and Universidad Autónoma de Chile approved the study and all subjects provided written informed consent (approval code: 063-15).
A venous blood sample (5 ml) was obtained from each participant by venipuncture and was stored in a serum Vacutainer tube (BD 366431). Samples were centrifugated at room temperature twice: at 800×g during 10min, followed by 10,000×g for 15min. The supernatant serum was recovered, placed into Eppendorf tubes, and stored at −80°C until RNA purification. For total RNA extraction, we used phenolchloroform separation and the miRNeasy Serum/Plasma kit (Cat. no 1071073, Qiagen, Venlo, The Netherlands) on a QIAcube (Qiagen). During the RNA extraction step, Glycogen (Cat. no AM9510, Invitrogen, Waltham, Massachusetts, USA) was used as a carrier. Quantification and integrity assessment of RNA was performed by Nanodrop spectrometry and on Agilent's Bioanalyzer. The eluate was concentrated using Ampure beads XP (Agencourt Bioscience, Beverly, Massachusetts, USA). A next-generation high-throughput sequencing platform (HiSeq2500, Illumina, San Diego, California, USA) optimized for low-input RNA (BGI Genomics, Shenzhen, Guangdong, China) was used for generating miRNA profiles. In order to ensure high quality data, we added NEBNext library kits, batch controls and internal small-noncoding-RNA spike-in controls (C. elegans miR-39). Sequencing libraries were indexed, and 10 samples were sequenced per lane, yielding 15 million sequences per sample on average. After initially trimming the RNAseq reads for adapters using AdapterRemoval (v2.1.7), we mapped the collapsed reads (generated by FASTX v0.14) to the human genome (hg38) using Bowtie2 (10 alignments per read were allowed). All multi-mapped reads with equivalent mapping score were counted. Finally, we compiled a comprehensive annotation set from miRBase (v21), using SeqBuster (v3.1) to get miRNA profiles and HTSeq (v0.7.2) to count the mapped reads.
Age and sex were similar in CS- and BS-COPD, but BMI was higher and school years significantly lower in the latter group (Table 1). Both smoking history and cumulative exposure to BS were high in the respective groups. Symptoms (mMRC, CAT and BODE), lung function tests and the 6MWT were similar in both groups. BS-COPD patients reported more exacerbations in the previous year than CS-COPD patients, although the difference was not significative. Finally, GOLD group distribution was similar in CS- and BS-COPD.
Demographic and Clinical Characteristics of Ever-smokers With COPD (CS) and Never-smokers With COPD Associated to Biomass Smoke (BS).
| CS-COPD n=15 | BS-COPD n=15 | P-Value | |
|---|---|---|---|
| Sex, M/F | 5/10 | 5/10 | |
| Age, years | 70.93±3.86 | 70.13±6.17 | .67 |
| BMI, kg/m2 | 26.94±4.77 | 30.12±6.92 | .15 |
| Scholarship, years | 9.87±3.92 | 4.06±3.31 | <.0001 |
| Smoking history, pack-years | 60.58±29.73 | –– | |
| Biomass smoke exposure, hour-years | –– | 316.67±223.47 | |
| Exacerbations in the last year | 1.73±1.44 | 2.93±1.69 | .52 |
| FEV1, % pred | 49.40±17.62 | 56.13±13.49 | .25 |
| FEV1/FVC, % | 56.40±11.02 | 58.06±7.58 | .63 |
| DLCO, % pred | 76.42±34.87 | 78.25±26.85 | .52 |
| Oxygen saturation, % | 94.38±3.74 | 91.42±4.65 | .29 |
| 6MWT, m | 360.75±124.85 | 250.00±144.39 | .25 |
| mMRC | 2.87±1.19 | 2.27±1.09 | .25 |
| CAT | 15.40±4.79 | 15.80±7.78 | .86 |
| BODE | 3.13±2.23 | 4.14±1.86 | .81 |
Definition of abbreviations: BMI body-mass index, FEV1 forced expiratory volume in 1s, FVC forced vital capacity, DLCO carbon monoxide diffusing capacity, 6MWT 6minutes walking test, mMRC modified Medical Research Council scale, CAT COPD assessment test, BODE body-mass, airflow Obstruction, Dyspnea and Exercise index.
Data presented as mean±standard deviation, unless otherwise indicated. Pulmonary function was assessed in all participants using standard procedures and equipment (Masterlab; Jaeger, Würzburg, Germany). Oxygen saturation was measured by pulse-oximetry (Ohmeda TuffSat, Soma Technology, Connecticut, USA). Dyspnea was measured using the modified Medical Research Council dyspnea scale (mMRC) and the COPD assessment test (CAT) was used to measure the impact of COPD on patient quality of life. The 6-minute walking test (6MWT) was also performed to each participant, according to European Respiratory Society/American Thoracic Society guidelines. The Body-mass, airflow Obstruction, Dyspnea and Exercise (BODE) score was calculated for each patient. Subjects were excluded if there was history of asthma, rhinitis or any extra-pulmonary disease affecting lung function, positive bronchodilator test, FEV1 increasing by ≥12% and 200ml and exacerbation or hospitalization record during the previous two months.
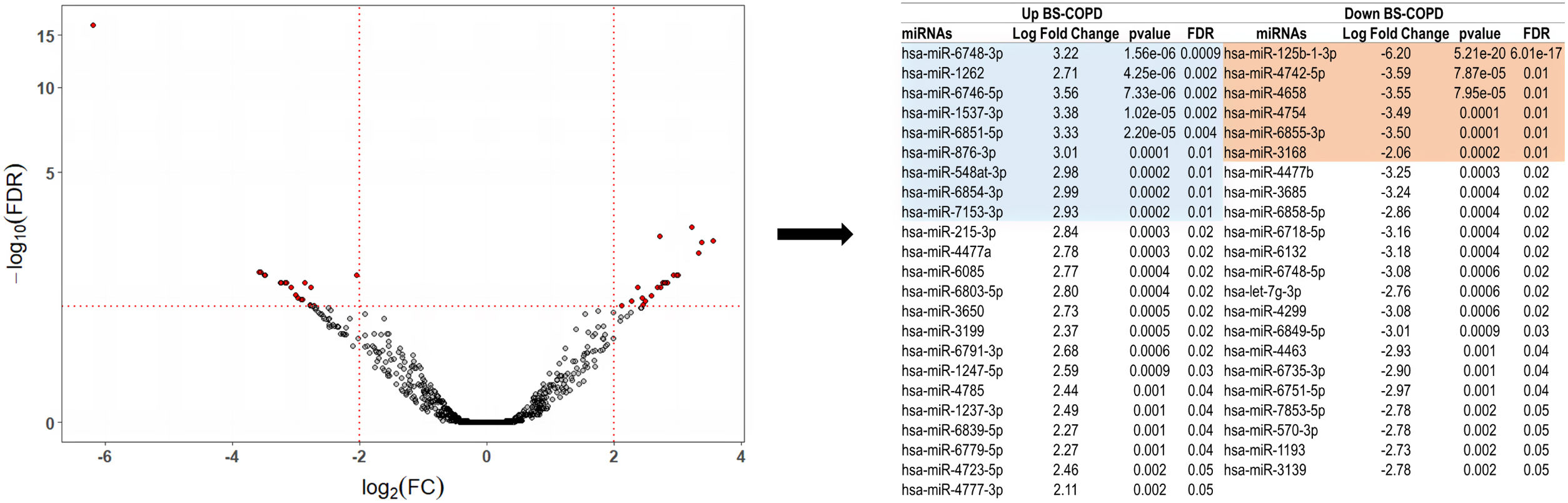
MicroRNAs expression was compared between groups using the R package edgeR. Forty-five miRNAs were differentially expressed in BS- vs. CS-COPD (|log2 fold change (FC)| of>2 and Benjamini–Hochberg false discovery rate (FDR)-correct P values <.05) (Fig. 1), 23 up and 22 down-regulated in the BS-COPD group. Among the miRNAs differentially expressed in BS-COPD, those with a false discovery rate (FDR)≤0.01 (9 upregulated and 6 downregulated) were screened with the microT-CDS algorithm (miRNA target genes score≥0.95).8 The analysis identified 287 genes related to the upregulated miRNAs and 173 genes related to the downregulated miRNAs (Fig. S1). The biological processes related to these genes were identified through Gene Ontology (GO) analysis, using the ClusterProfiler package. Eighteen terms were found to be involved in the differentially expressed miRNAs enrichment analysis (Fig. S2). Overall, these were related to four main processes: cell responses to hypoxia, neuronal development and differentiation, regulation of cell cycle G2/M phase transition, and chromatin and histone modification. In the enrichment map showed in Fig. S3, highly similar terms were gathered into clusters that underpin general trends. Terms related to cell responses to hypoxia and regulation of cell cycle G2/M phase transition had intersecting gene sets with each other. We also used X2K to construct the upstream regulatory network of the differentially expressed miRNAs.9 This analysis revealed that CREB1, PPARG, YY1, CHD1, UBTF, SPI1, TAF1, and REST were the most important transcription factors (TFs) regulating gene expression, whereas GSK3B, MAPK14, CDK1, HIPK2, CK2ALPHA, ERK2, MAPK3 and ERK1 were the most important kinases controlling the expression of genes (Fig. S4).
Differential miRNA expression in never-smoking COPD patients exposed to biomass smoke versus cigarette smoking COPD patients. (A) Volcano plot showing the differential miRNA expression (in fold change on x-axis) and significance level (−log10-FDR value on y-axis). To exclude poorly detectable miRNAs, data was pre-processed so that total counts per million (CPM) mapped reads for each miRNA in all samples pertaining to the comparison subset was>10. After that, the filtered data were normalized using the trimmed mean of M-values (TMM) normalization method in EdgeR. Of the 2609 miRNAs analyzed, 1460 were discarded because they were poorly detectable according to the quality criteria.
A growing body of evidence have shown up/down-regulation of several miRNAs in CS-COPD patients when compared to subjects without COPD.10–13 Interestingly, none of the miRNA described so far were found to be differentially expressed between CS-COPD and BS-COPD in our study. Moreover, there is a paucity of data regarding the effects of BS on miRNA expression.14
Regarding the TFs related to the differentially expressed miRNAs, our analysis identified PPARG, YY1 and CREB1 as those with the highest interactions with other genes. Interestingly, alveolar hypoxia has been shown to lead to selective phosphorylation and activation of CREB1 in the lungs, suggesting an important role of this TF in the pulmonary response to decreased oxygen levels.15 With respect to the top predicted kinases of the inferred upstream regulatory network, GSK3β was enriched more than the rest. Glycogen synthase kinase-3β (GSK3β) is regulated by oxidative stress and may play a key role in oxidant-mediated signaling during COPD inflammation. Moreover, a recent miRNAs study developed in bronchial biopsies of COPD patients pointed to GSK3β as potential regulator of chronic mucus hypersecretion.16 Remarkably, BS exposure has been related to cough and mucus overproduction,17 and BS-COPD patients have more cough and phlegm symptoms than CS-COPD subjects.18
Several limitations in our design should be kept in mind, which precludes making solid conclusions. These include that our study lacks of a replication cohort, being a hypothetical work based on in silico data pending of confirmation with functional studies. In addition, the sample size of our cohort was relatively small (n=30) and we did not include a group of control subjects without COPD. Nevertheless, the high number of miRNAs studied support our findings, which may be useful in order to set specific therapeutic targets or prognostic indicators for future investigations on COPD related to BS exposure.
The results of our study show that the expression of circulating miRNAs is different in CS- and BS-COPD patients. Since differentially expressed miRNAs in BS-COPD relate to genes and pathways involved in hypoxemia and mucus hypersecretion, we hypothesize that these mechanisms are more relevant in these patients.
Authors’ ContributionsStudy concept and design: JO; Data acquisition: RSS, JO; Data analysis: JO, RDP; Data interpretation: JO, RDP, HDH, AA; Funding acquisition: JO; Investigation: JO, RDP; Methodology: JO, RDP; Supervision: JO; Writing – original draft: JO; Writing – review & editing: JO, RDP, HDH, RSS, AA.
Statement of EthicsThis research complies with the guidelines for human studies and was conducted ethically in accordance with the World Medical Association Declaration of Helsinki.
Funding SourcesFunding support for this study was provided by the Chilean National Science and Technology Fund (CONICYT), FONDECYT Project N° 11150022 (JO).
Conflicts of InterestThe authors declare no conflicts of interest.
Authors thank participants in the study for their willingness to contribute to medical research, Ms. Viviana Parra, Ms. Hanuxa Celedón and Mr. Hans Jürgen Kürsch for their technical assistance and the American Thoracic Society (ATS) Methods in Epidemiologic, Clinical and Operations Research (MECOR) Program for its support with the study's concept.