Asthma inflammation may feature an imbalance between oxidative stress and antioxidant defenses. Oxidative stress induces propagation of airways inflammation and corticosteroid insensitivity contributing to poor asthma control, and frequent severe acute exacerbations. This study assessed inflammation and oxidative stress in severe asthmatic subjects and evaluated the possible correlations between inflammatory and oxidative stress markers investigated and asthma severity.
Material and methodFifty-three patients with severe asthma, 11 patients with mild-moderate asthma and 12 healthy subjects were enrolled and underwent fractional exhaled nitric oxide (FENO) analysis and blood and sputum count cell collection. The content of mitochondrial DNA (MtDNA) and nuclear DNA (nDNA) was measured in exhaled breath condensate (EBC) by Real Time PCR and the ratio between MtDNA/nDNA was calculated. We detected MtDNA/nDNA in the EBC of severe asthmatics.
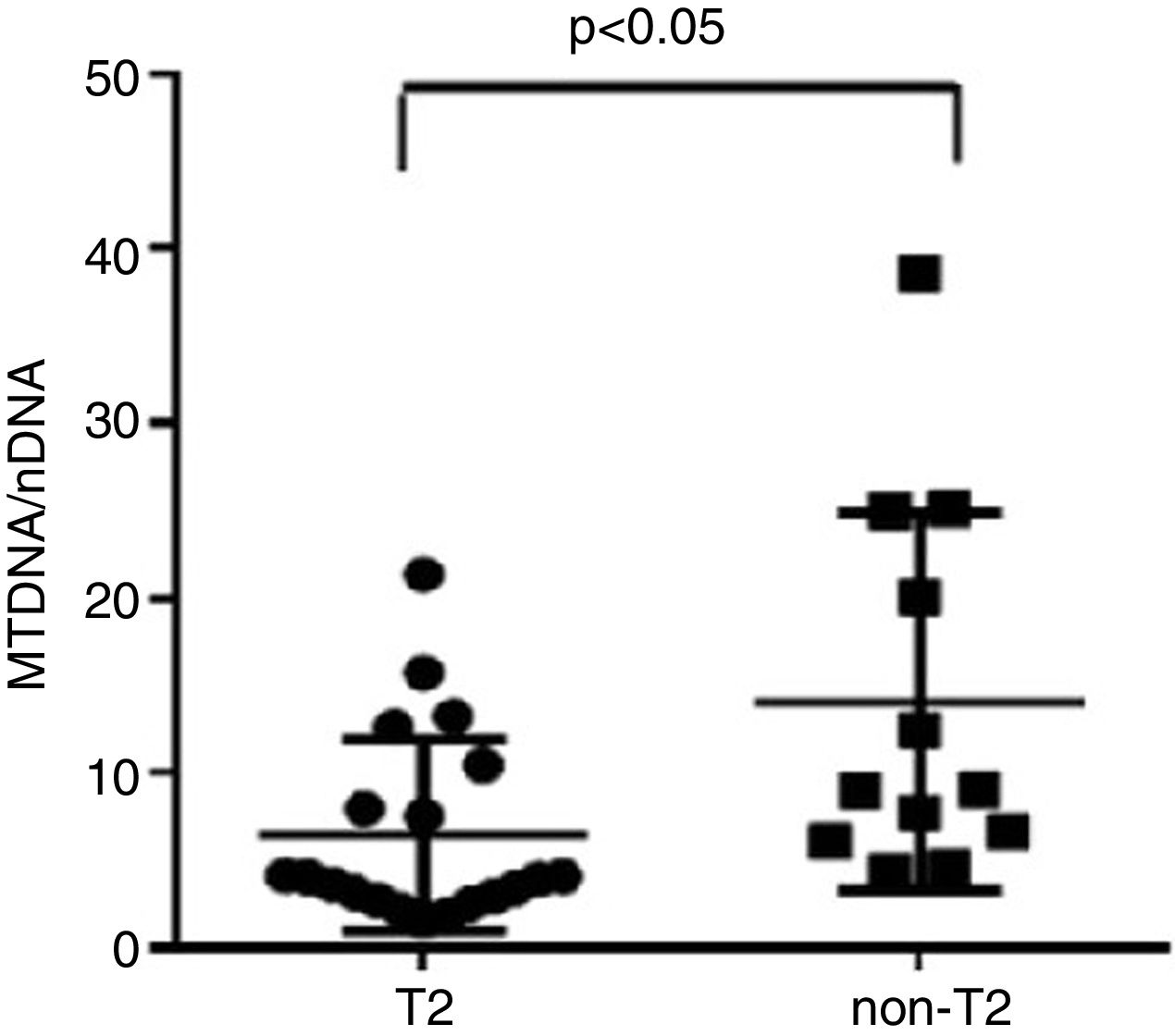
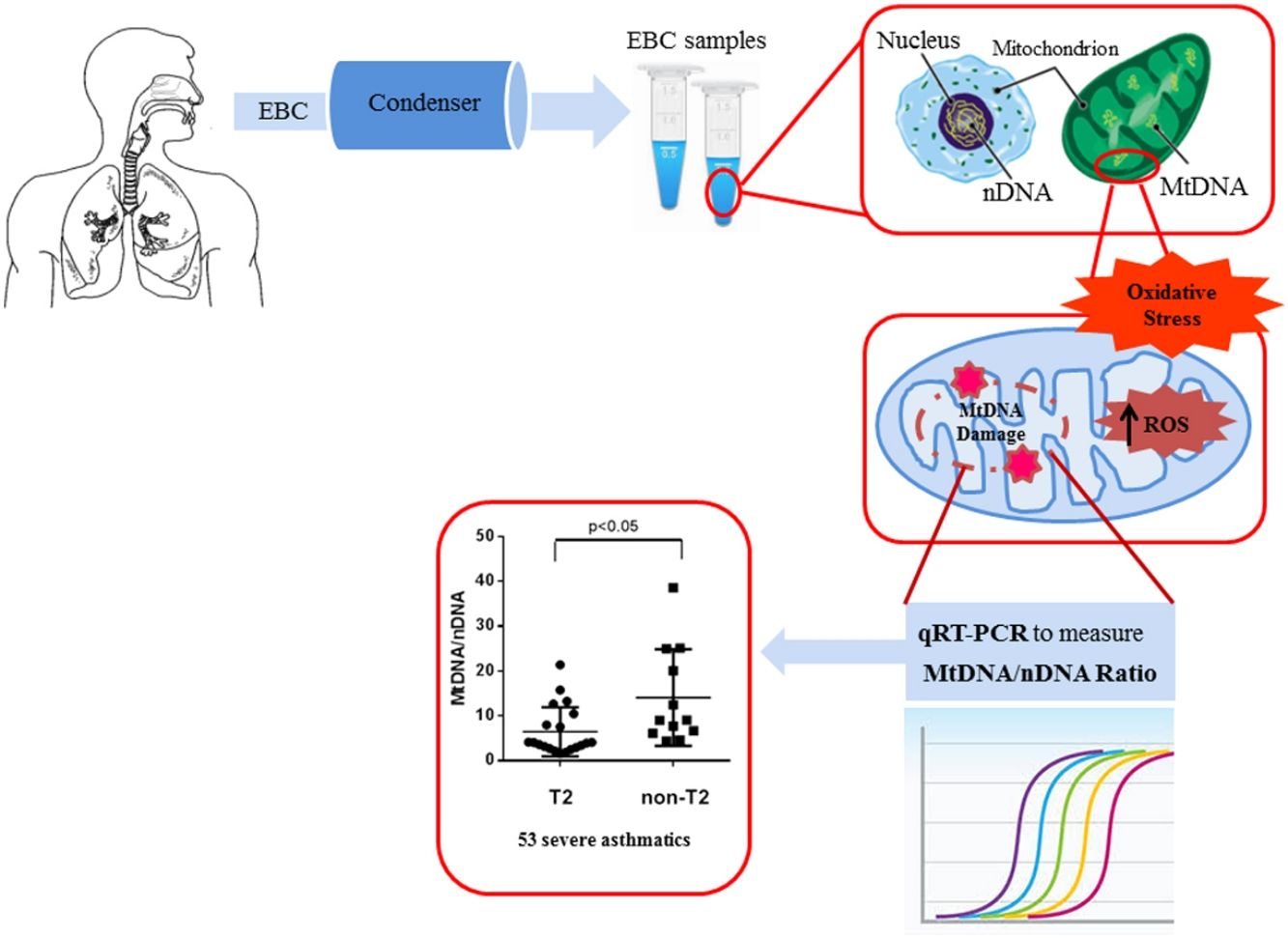
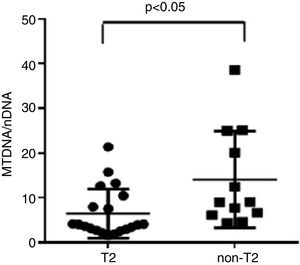
ResultsWe found higher exhaled MtDNA/nDNA in severe asthmatics respectively compared to mild-moderate ones and to healthy controls (10.4±2.2 vs 7.9±2.5, p<0.05 and 10.4±2.2 vs 6.51±0.21, p<0.05). The level of exhaled MtDNA/nDNA was significantly higher in Non-T2 endotype severe asthmatics than T2 (14.07±10. 8 vs 6.5±5.5, p<0.05).
ConclusionOxidative stress marker (MtDNA/nDNA) is increased significantly with asthma severity and may be useful for endotyping severe asthma.
La inflamación en el asma puede presentar un desequilibrio entre el estrés oxidativo y las defensas antioxidantes. El estrés oxidativo induce la propagación de la inflamación de las vías aéreas y la insensibilidad a los corticosteroides, lo que contribuye a un control deficiente del asma y a frecuentes exacerbaciones agudas graves. Este estudio evaluó la inflamación y el estrés oxidativo en sujetos asmáticos graves y estudió las posibles correlaciones entre los marcadores de estrés inflamatorio y oxidativo investigados y la gravedad del asma.
Material y métodoSe incluyó a 53 pacientes con asma grave, 11 pacientes con asma leve a moderada y 12 sujetos sanos, a los que se les realizó un análisis de fracción exhalada de óxido nítrico (FeNO) y un recuento celular del esputo y de sangre. Se midió el contenido de ADN mitocondrial (ADNmt) y ADN nuclear (ADNn) en el condensado de aire exhalado (CAE) mediante PCR en tiempo real y se calculó la ratio ADNmt/ADNn. Detectamos ADNmt/ADNn en el CAE de los asmáticos graves.
ResultadosEncontramos unos niveles más altos de ADNmt/ADNn exhalados en los asmáticos graves en comparación con los leves moderados y los controles sanos (respectivamente, 10,4±2,2 frente a 7,9±2,5, p<0,05 y 10,4±2,2 frente a 6,51±0,21, p<0,05). El nivel de ADNmt/ADNn exhalado fue significativamente mayor en los asmáticos graves de endotipo no-T2 que en los T2 (14,07±10,8 frente a 6,5±5,5, p<0,05).
ConclusiónEl marcador de estrés oxidativo (ADNmt/ADNn) aumenta significativamente con la gravedad del asma y puede ser útil para endotipar el asma grave.
Asthma is a complex, heterogeneous, chronic respiratory disease with a wide clinical spectrum.1 Analytical clustering methods have revealed phenotypes that include dependence on high-dose corticosteroid treatment, severe airflow obstruction and recurrent exacerbations associated with an allergic background and late onset of disease.2 Blood and sputum eosinophilia have been used to distinguish patients with high T helper lymphocyte type-2 type (T2)-inflammation and to predict therapeutic response to treatments targeted towards T2-associated cytokines (endotyping).3,4 In this direction, a new run in the validation of recognized inflammatory markers and in the discovery of new biomarkers characterize this era. The keyword is non-invasiveness, especially when we speak of patients affected by severe asthma.2
Several studies have been directed to the immunologic genesis of asthmatic airways inflammation, but recent research paid attention to the most inflammatory effector cells involved in the production of reactive oxygen species (ROS) and reactive nitrogen species (RNS) which produce pathophysiologic outcomes of airways reactivity, injury and remodelling.5 Oxidative stress induces propagation of airways inflammation and corticosteroid insensitivity6 and is associated with poor asthma control, and frequent severe acute exacerbation. Although the knowledge of the link between airways inflammation and oxidative stress in asthma is well known,7 it still remains unclear whether oxidative stress is a distinct clinical and pathological feature in asthma. This could be the reason why a new focus of severe asthma research is now directed towards the analysis of oxidative stress markers.8
An increased interest is, today, directed towards the mitochondrial cellular function beyond energy production.9 Malfunctioning mitochondria change the bioenergetics of the cell and its metabolic profile to favour systemic inflammation that drives asthma.10 According to this, the measurement of mitochondrial DNA (MtDNA) through the mitochondrial genome versus nuclear genome ratio, termed Mt/N, as new intriguing oxidative stress marker is gaining ever greater importance.11 In our previous study, we assessed oxidative stress in human plasma by measuring the levels of Reactive Oxygen Metabolites (ROMs) using the d-ROM test, and then we showed that MtDNA/nDNA ratio correlates with these markers of oxidative stress.12
Our group recently showed an increase of MtDNA/nDNA in patients with asthma, COPD and ACOS, measuring it for the first time in the EBC, a sample from airways that is collectable with complete non-invasiveness by just breathing at tidal volume in the condenser device.13
The aim of this preliminary study was to analyze biomarkers of airways inflammation already validated for phenotyping severe asthma and a new marker of oxidative-stress, the MtDNA/nDNA in the EBC. We furthermore wanted to evaluate the potential role of oxidative-stress in endotyping severe asthma and therefore to investigate the possible correlations between MtDNA/nDNA and clinical data and inflammatory biomarkers.
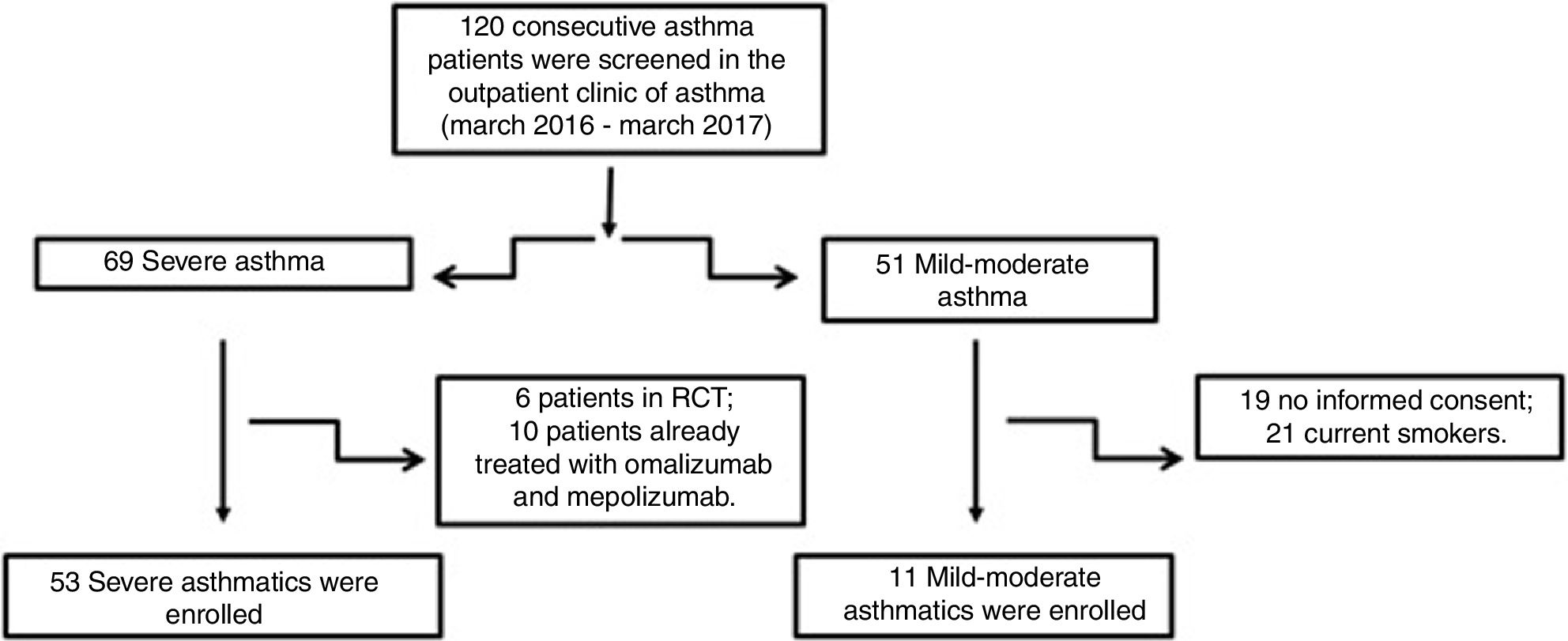
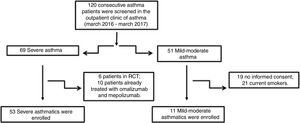
Material and methodsPatientsFifty-three patients with severe asthma (SA) were consecutively enrolled according to the international ERS/ATS guidelines14 during a period of 12 months. Patients were recruited for the study from the outpatient clinic of severe asthma of the Institute of Respiratory Diseases of the University of Foggia, Italy from March 2016 to March 2017. Patients with SA who were enrolled in randomized controlled trials and patients already treated with omalizumab and mepolizumab were excluded.
Patients with other comorbidities were excluded. We also enrolled 11 patients with controlled persistent mild-moderate asthma (MA) and 12 healthy subjects (HS) as controls. All subjects enrolled were non- or former smokers for at least 1 year and were assessed at a period of stability and at least four weeks after an upper respiratory tract infection.
All subjects filled out a questionnaire about information on their medical history, then they underwent a spirometry, physical examination and atopy assessment. The second time, the EBC was collected from all patients and a blood analysis for eosinophils, FENO measurement and sputum induction was performed.
This study was conducted in accordance with the amended Declaration of Helsinki. Written informed consent was obtained from all subjects and the study was approved by the institutional ethics committee of the University of Foggia (institutional review board approval number 17/CE/2014).
QuestionnairesThe clinical evaluation of patients enrolled in the study consisted of the administration of the most important validated questionnaires to assess symptoms control (ACT, ACQ),15 quality of life (AQLQ-S).16
Atopic statusSkin prick test (SPT) was performed for a panel of inhalant allergens as previously described for common aeroallergens (Lofarma, Italy).17
Lung functionForced spirometric (before and after bronchodilation) and plethysmographic lung volumes (Sensormedics, USA) were determined following international standards in all patients.18
Blood collectionVenous blood samples were drawn and put into a tube containing EDTA. The count of blood cells was determined using an automated analyzer. The total number of white blood cells was multiplied by the percentage of eosinophils to provide the absolute eosinophil count (eosinophils×109/L).
Induced sputum collection and processingAccording to the method described by Spanevello et al., in healthy subjects and in mild-moderate asthmatics, sputum was induced through inhalation of hypertonic saline solution (4.5%) with an ultrasonic nebulizer (DeVilbiss 65; DeVilbiss Corporation,Somerset, PA) and analyzed after selection of plug.19 In severe asthmatic subjects we used the spontaneous sputum or the sputum produced after inhalation of isotonic saline solution. The sputum (spontaneous or induced) was used for cytological analysis.
Measurement of FENOThe Medisoft FENO+device, which is a semiportable for repeatable multi-flow measurement of exhaled NO with offline measurement, was used. It has a software package that provides step-by-step online quality control. The measurement range is 0–600ppb. FENO was measured using a previously described restricted breath technique, which employed expiratory resistance and positive mouth pressure to close the velum and exclude nasal NO: expiratory flow measurements at 50mL/s and a 350mL/s have been evaluated. Repeated exhalations were performed until three plateaus agreed within 5%.20,21
EBC and DNA extractionOne (1) ml of EBC was collected in one sitting from each patient at the time of diagnosis, by using a condenser, which allowed for the non-invasive collection of non-gaseous components of the expiratory air (EcoScreen Jaeger, Wurzburg, Germany). The condensate was collected on ice at −20°C, transferred to 1.5ml polypropylene tubes, and immediately stored at −70°C for subsequent analysis. We used the condensate within one month from storage. The collected EBC was concentrated with Microcon-30kDa Centrifugal Filter Unit with Ultracel-30 membrane (Merk Millipore, Billerica, MA, USA) and directly measured by quantitative real time PCR method using an Applied Biosystems 7300 real-time PCR System.22 The DNA concentration adjusted to 10ng/μl refers to DNA extracted from blood. The Real-Time PCR was performed by TaqMan® Universal PCR Master Mix (PE Applied Biosystems).
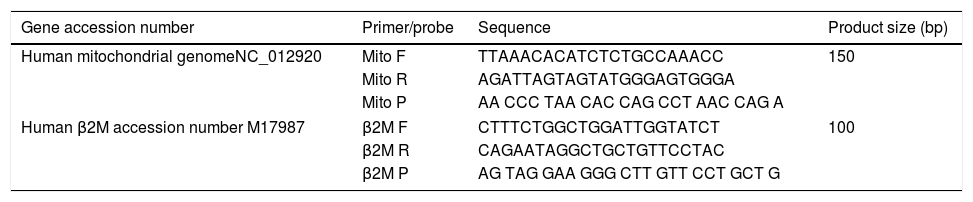
Quantitative real-time PCRMitochondrial DNA content was measured by quantitative real time PCR (qPCR) method using an Applied Biosystems 7300 real-time PCR System (PE Applied Biosystems). MtDNA was measured by quantification of a unique mitochondrial fragment relative to a single copy region of beta-2-microglobulin nuclear gene (β2M).23 Primers, Probes (IDT, Integrated DNA Technologies, USA) and gene accession numbers are listed in Table 1. Mitochondrial DNA and β2M probes were labelled at 5′end with 6 FAM and MAX fluorescent dyes respectively and both probes contained BHQ-1 as a quencher dye at 3′end. The PCR mix was: 1× TaqMan® Universal PCR Master Mix (PE Applied Biosystems), 200nM of each primer, 125nM of TaqMan Probe, 50ng of total genomic DNA extract in a 20μl PCR reaction. Quantitative real-time PCR conditions were 2min at 50°C and 10min at 95°C, followed by 40 cycles of 15s of denaturation at 95°C and 60s of annealing/extension at 60°C. The positive controls (extracted from normal healthy persons) and negative control (DDW+master mix) were added for every PCR run. Standard curves obtained from serial dilutions of PCR-amplified target sequences were used for the quantification of MtDNA and nuclear genome, and then the ratio of MtDNA/nDNA was calculated.
Primers/probes sequences for MtDNA/nDNA determination using real time qPCR; mitochondrial DNA and β2M probes were labelled at 5′end with 6 FAM and MAX fluorescent dyes respectively, both probes contained BHQ-1 as a quencher dye at 3′end.
| Gene accession number | Primer/probe | Sequence | Product size (bp) |
|---|---|---|---|
| Human mitochondrial genomeNC_012920 | Mito F | TTAAACACATCTCTGCCAAACC | 150 |
| Mito R | AGATTAGTAGTATGGGAGTGGGA | ||
| Mito P | AA CCC TAA CAC CAG CCT AAC CAG A | ||
| Human β2M accession number M17987 | β2M F | CTTTCTGGCTGGATTGGTATCT | 100 |
| β2M R | CAGAATAGGCTGCTGTTCCTAC | ||
| β2M P | AG TAG GAA GGG CTT GTT CCT GCT G | ||
Descriptive statistics (i.e. means, standard deviations, percentages) were applied to summarize the continuous and categorical variables. Continuous variables were analyzed by ANOVA, categorical variables by χ2 tests. Correlations between MtDNA/nDNA and clinical, functional and inflammatory data were assessed using the Pearson correlation coefficient (SPSS 22.0, INC, Chicago, III). p value<0.05 was considered significant.
ResultsFig. 1 presents the consort diagram of the study, and Table 2 the main demographic, clinical and functional characteristics of participants included in the analysis.
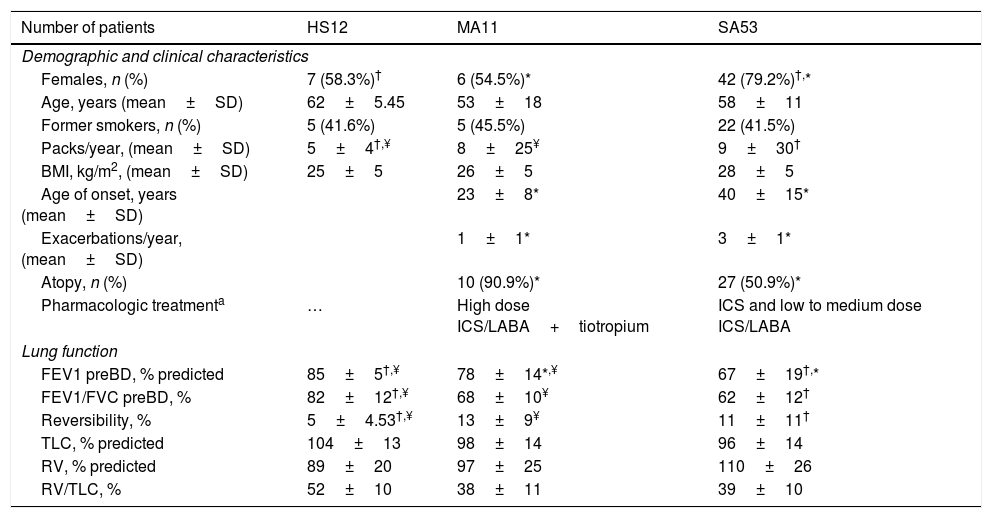
Demographic, clinical and functional characteristics of study population.
| Number of patients | HS12 | MA11 | SA53 |
|---|---|---|---|
| Demographic and clinical characteristics | |||
| Females, n (%) | 7 (58.3%)† | 6 (54.5%)* | 42 (79.2%)†,* |
| Age, years (mean±SD) | 62±5.45 | 53±18 | 58±11 |
| Former smokers, n (%) | 5 (41.6%) | 5 (45.5%) | 22 (41.5%) |
| Packs/year, (mean±SD) | 5±4†,¥ | 8±25¥ | 9±30† |
| BMI, kg/m2, (mean±SD) | 25±5 | 26±5 | 28±5 |
| Age of onset, years (mean±SD) | 23±8* | 40±15* | |
| Exacerbations/year, (mean±SD) | 1±1* | 3±1* | |
| Atopy, n (%) | 10 (90.9%)* | 27 (50.9%)* | |
| Pharmacologic treatmenta | … | High dose ICS/LABA+tiotropium | ICS and low to medium dose ICS/LABA |
| Lung function | |||
| FEV1 preBD, % predicted | 85±5†,¥ | 78±14*,¥ | 67±19†,* |
| FEV1/FVC preBD, % | 82±12†,¥ | 68±10¥ | 62±12† |
| Reversibility, % | 5±4.53†,¥ | 13±9¥ | 11±11† |
| TLC, % predicted | 104±13 | 98±14 | 96±14 |
| RV, % predicted | 89±20 | 97±25 | 110±26 |
| RV/TLC, % | 52±10 | 38±11 | 39±10 |
Abbreviations. HS: healthy subjects; SA: severe asthma; MA: mild-moderate asthma; BMI: body mass index; FEV1: forced expiratory volume; FEV1/FVC: forced expiratory volume/forced vital capacity; BD: bronchodilator; TLC: total lung capacity: RV: residual volume; RV/TLC: residual volume/total lung capacity.
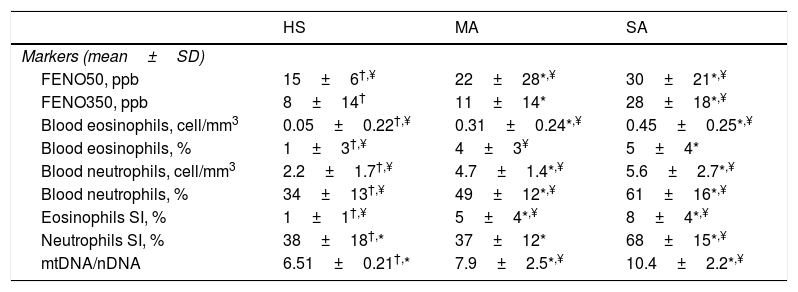
We evaluated non-invasive inflammatory biomarkers and a new oxidative stress marker that is MtDNA/nDNA in EBC (Table 3). Severe asthmatics of our study presented significantly higher FENO50 and FENO350 and higher blood and sputum eosinophils than mild-moderate ones and healthy controls.
Inflammatory and oxidative-stress markers.
| HS | MA | SA | |
|---|---|---|---|
| Markers (mean±SD) | |||
| FENO50, ppb | 15±6†,¥ | 22±28*,¥ | 30±21*,¥ |
| FENO350, ppb | 8±14† | 11±14* | 28±18*,¥ |
| Blood eosinophils, cell/mm3 | 0.05±0.22†,¥ | 0.31±0.24*,¥ | 0.45±0.25*,¥ |
| Blood eosinophils, % | 1±3†,¥ | 4±3¥ | 5±4* |
| Blood neutrophils, cell/mm3 | 2.2±1.7†,¥ | 4.7±1.4*,¥ | 5.6±2.7*,¥ |
| Blood neutrophils, % | 34±13†,¥ | 49±12*,¥ | 61±16*,¥ |
| Eosinophils SI, % | 1±1†,¥ | 5±4*,¥ | 8±4*,¥ |
| Neutrophils SI, % | 38±18†,* | 37±12* | 68±15*,¥ |
| mtDNA/nDNA | 6.51±0.21†,* | 7.9±2.5*,¥ | 10.4±2.2*,¥ |
Abbreviations. HS:healthy subjects; SA: severe asthma; MA: mild-moderate asthma; FENO: fractional exhaled nitric oxide; mtDNA/nDNA: mitocochondiral DNA/nuclear genome DNA; SI: induced sputum.
MtDNA/nDNA was detectable in all EBC samples but, because of its real-life nature, three healthy controls, two mild-moderate asthmatics and seven severe asthmatics didn’t consent to the execution of the procedure. We found higher MtDNA/nDNA in EBC of patients with SA than with MA (10.4±2.2 vs 7.9±2.5, p<0.05).
Severe asthmatics were divided in two groups according to eosinophil count in blood and sputum samples: T2 (25 SA patients), manifested by elevated sputum>3% and/or blood eosinophils ≥400cells/μl on at least two occasions, and non-T2 (21 SA patients), with elevated sputum neutrophils ≥76% but blood eosinophils<300cells/μl on at least two occasions (Table 4).
Severe asthmatic endotypes.
| T225 | Non-T221 | |
|---|---|---|
| Markers, (mean±SD) | ||
| FENO50, ppb | 33±18* | 22±17* |
| FENO350, ppb | 23±23* | 14±21* |
| Blood eosinophils, cell/mm3 | 0.45±0.28* | 0.19±0.25* |
| Blood neutrophils, cell/mm3 | 3.8±0.32* | 8.6±2.7* |
| Eosinophils SI, % | 8±2* | 2±1* |
| Neutrophils SI, % | 38±18+ | 88±15* |
| mtDNA/nDNA | 6.5±5.5* | 14.07±10.8* |
Abbreviations. T2: severe asthma T-helper2; Non-T2: severe asthma non T-helper2; FENO: fractional exhaled nitric oxide; mtDNA/nDNA: mitocochondiral DNA/nuclear genome DNA; SI: induced sputum.
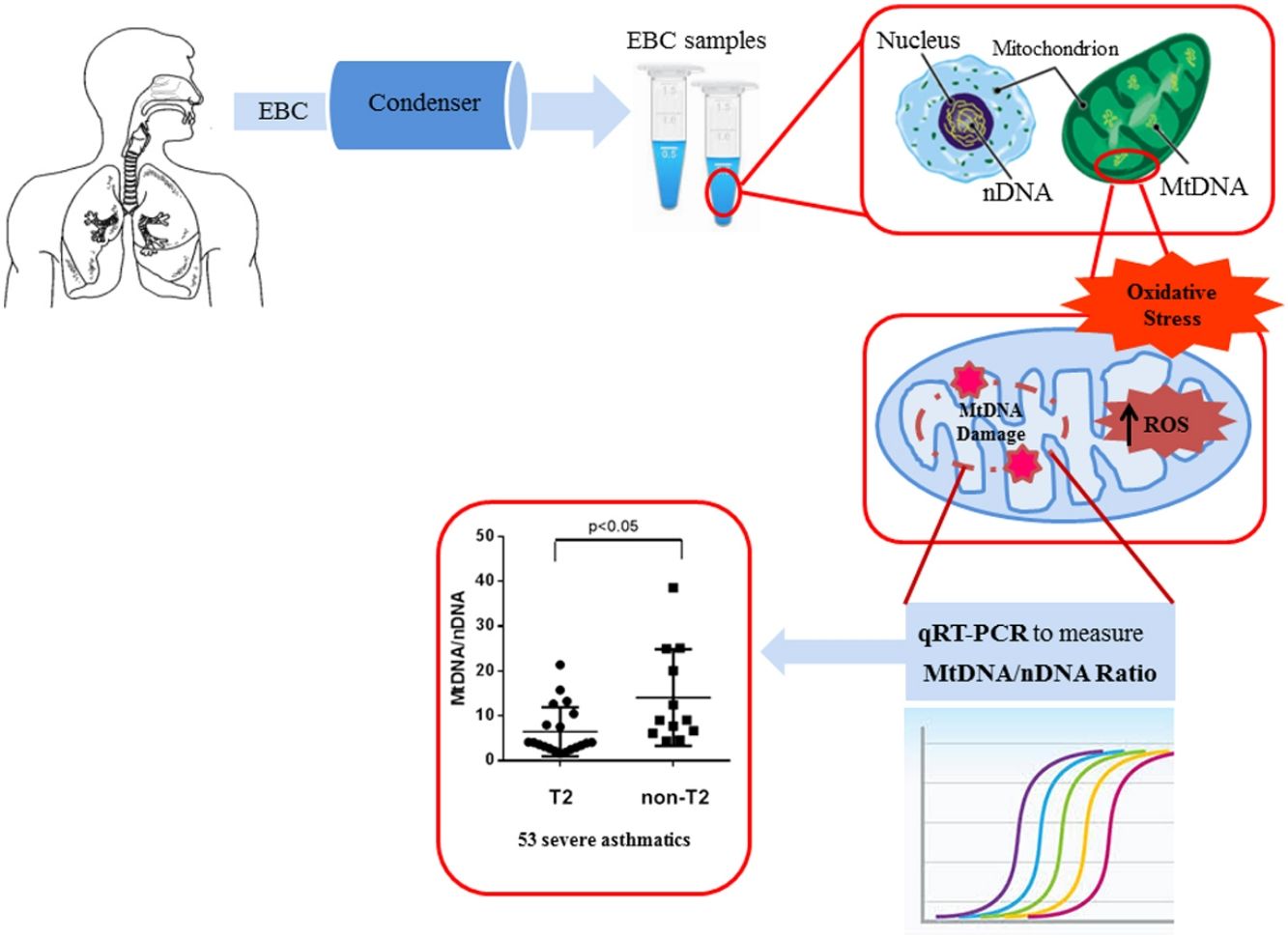
We observed that non-T2 endotype group showed a significantly higher exhaled MtDNA/nDNA than T2 endotype (14.07±10.8 vs 6.5±5.5, p<0.05) (Fig. 2).
In addition, Mt/N values had no significant correlation with pulmonary function tests. No other correlations between MtDNA/nDNA and clinical data were found.
DiscussionIn this work, the main result is the increased inflammation and oxidative stress resulting in an alteration of the amount of MtDNA, higher in the EBC of severe asthmatic patients compared to those with mild-moderate asthma. A significantly higher concentration was observed in severe asthmatics of non-T2 endotype.
In consideration of the importance that is directed towards a personalized clinical approach to severe asthma, in this study we not only enrolled severe asthmatic subjects but we also tried to divide them according to the endotype.3 Accordingly with what was widely written in previous years, we found that different endotypes were biologically recognizable: T2 endotype showed an eosinophilic pattern characterized by higher FENO and blood and sputum eosinophils; on the contrary non-T2 endotype showed higher sputum neutrophils. But the stability and the natural history of these biological phenotypes are poorly understood.24 Kuo et al.25 reasoned that endotyping severe asthma using biological features may not only not provide a precise definition of severe asthma, but may even preclude the collection of adequate information on the underlying mechanisms of each phenotype.
However, endotypes can play a role in guiding treatment decisions.26
Oxidative stress is the imbalance between oxidants and antioxidants, resulting in favour of oxidants. The asthmatic airways are exposed both to several exogenous reactive oxygen species (ROS), such as cigarette smoke or air pollution, and endogenous ROS, mostly generated by inflammatory cells, such as eosinophils and neutrophils.27,28
ROS are highly reactive molecules, short-lived and difficult to measure; therefore, products of ROS reactions are measured instead to estimate the oxidative stress burden and inflammation.29
Different contributions on the measurement of changes in mitochondrial DNA (MtDNA) content in vitro, measured as ratio Mt/N using qPCR, are available today in several human diseases.30 Furthermore, MtDNA has been studied as an expression of oxidative stress in asthma, COPD, lung cancer and obstructive sleep apnoea, but it has been mainly investigated systemically, although the pathogenetic mechanisms begin in the airways and only later progress to systemic circulation.
Yang Ai et al.31 analyzed the D-loop region of exhaled MtDNA underline that it is possible to study mitochondrial DNA in this biological sample. Even our group, in previous studies, analyzed mitochondrial DNA balance in lung diseases such as OSAS and ACOS reporting an increase of MtDNA/nDNA in the blood and EBC of subjects affected by these pathologies.13,32
In this study we have shown, for the first time, that there is an increase of mitochondrial DNA, as an imbalance of Mt/N, in EBC from asthma patients.
Our hypothesis is that the presence of oxidative stress in severe asthmatics can induce an alteration of the transcriptional and replication machinery of mitochondrial biogenesis which would be up-regulated resulting in an increased mitochondrial activity. At this regard, our findings are perfectly in line with previous reports as that of Piotrowski et al. that showed a high concentration of 8-isoprostane33 and of Paredi et al. that reported elevated levels of methane in severe asthmatics.34 Furthermore, we observed that the highest concentration of the oxidative-stress marker was in non-T2 endotype. This result may have at least two justifications: the first may be the oxidative stress augments airway neutrophil recruitment and chemokine production in severe asthma.35 The second may be the former smoking habit of 15 subjects (71%) belonging to the non-T2 endotype; we know that the cigarette is itself an important source of oxidant.
In the majority of the previous studies, authors found a correlation between markers of oxidative-stress and measures of diagnosis and control and other inflammatory markers. In line with Bishopp et al. which reported an increase of the oxidative stress burden in severe asthma.29 In this study we tried to analyze the correlation between MtDNA/nDNA and clinical index of asthma as ACT and number of exacerbations and the results did not correlate. Previous papers highlighted the necessity to clarify whether oxidative status is associated with the degree of asthma.36 So we also analyzed possible correlations between MtDNA/nDNA and lung function without expected results. There is a strong link between oxidative stress and airways inflammation. We know that the amounts of ROS are largely responsible for the airway inflammation in asthma,37 promoting the expression of numerous pro-inflammatory cytokines such as tumour necrosis factor (TNF)-α, interleukin (IL-1, IL-6, and IL-8),38 which subsequently induce the activation of inflammatory cells in the respiratory tract.39 Interestingly, these inflammatory cells including macrophages, eosinophils, neutrophils, and monocytes have been shown to generate ROS themselves in order to kill the invading bacteria.40 Thus we also analyzed possible correlations between MtDNA/nDNA and FENO50 that is a recognized T2 eosinophilic inflammatory marker and correlates with asthma severity but we didn’t find expected results. We believe that it could be only due to the small number of samples of EBC because of its real-life nature, our database contains some missing data. This is a small preliminary study and our conclusion on the utility of MtDNA as an oxidative stress marker requires confirmation on larger studies. We agree that to compare MtDNA with other recognized markers of oxidative stress would improve its value and therefore we are planning a future study where we will compare MtDNA with other recognized markers of oxidative stress dosable in the EBC as the 8-isoprostane, the reactive oxygen species (ROS), the malondialdehyde (MDA) and the hydroperoxyl radical (HO2). Furthermore, we recognize as a limitation of our results that treatment with steroids and microbial dysregulation in SA could have possible confounding effects.
One strength of the present study lies on the use of three well characterized populations of individuals with asthma.
ConclusionIn conclusion, to our knowledge, this is the first time that an endotyping approach aimed at identifying differences in inflammatory and oxidative-stress markers between severe asthmatic patients is used. We believe that enriching severe asthma assessment and management with old and new markers, as those analyzed in this study, could allow for better endotyping of severe asthmatic patients and represent an ideal approach to improve patient outcomes.
Author agreementThe authors have seen and approved the final version of the manuscript. The authors warrant that the article is an original work, hasn’t received prior publication and isn’t under consideration for publication elsewhere.
Ethical approval and consent to participateThis study was conducted in accordance with the amended Declaration of Helsinki and the study was approved by the institutional ethics committee of the University of Foggia (institutional review board approval number 17/CE/2014).
Informed consentInformed consent was obtained from all individual participants included in the study.
Availability of data and materialsSource data and material will be made available upon reasonable request.
Authors’ contributionsGEC and GS designed the study; GS, GC, CQ, and PS contributed to the clinical and laboratory work for the study; GEC and GS drafted the article and revised it critically for important intellectual content; GEC, DL and MPFB contributed to final approval of the version to be published. All authors read and approved the final manuscript.
Funding sourceThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interestThe authors declare that they have no conflict of interest.