Identification of suitable biomarkers that facilitate the screening and evaluation of pediatric obstructive sleep apnea (OSA) and its severity was explored.
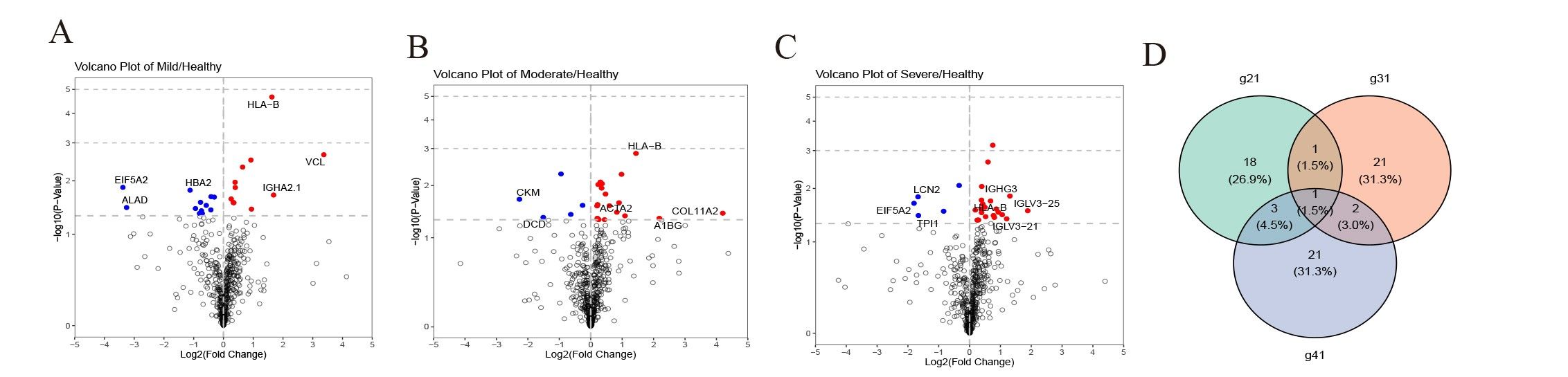
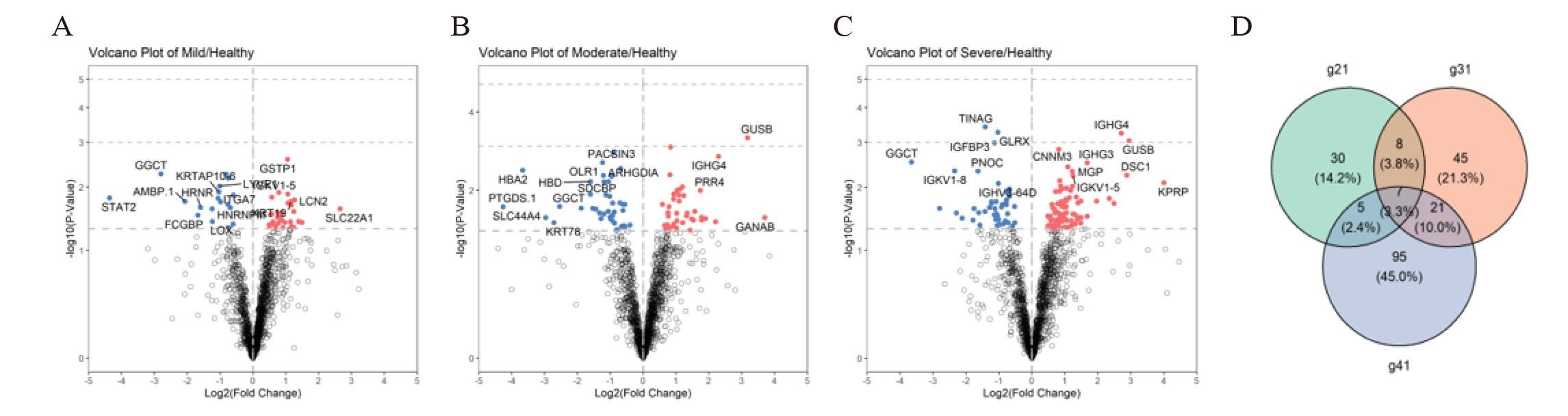
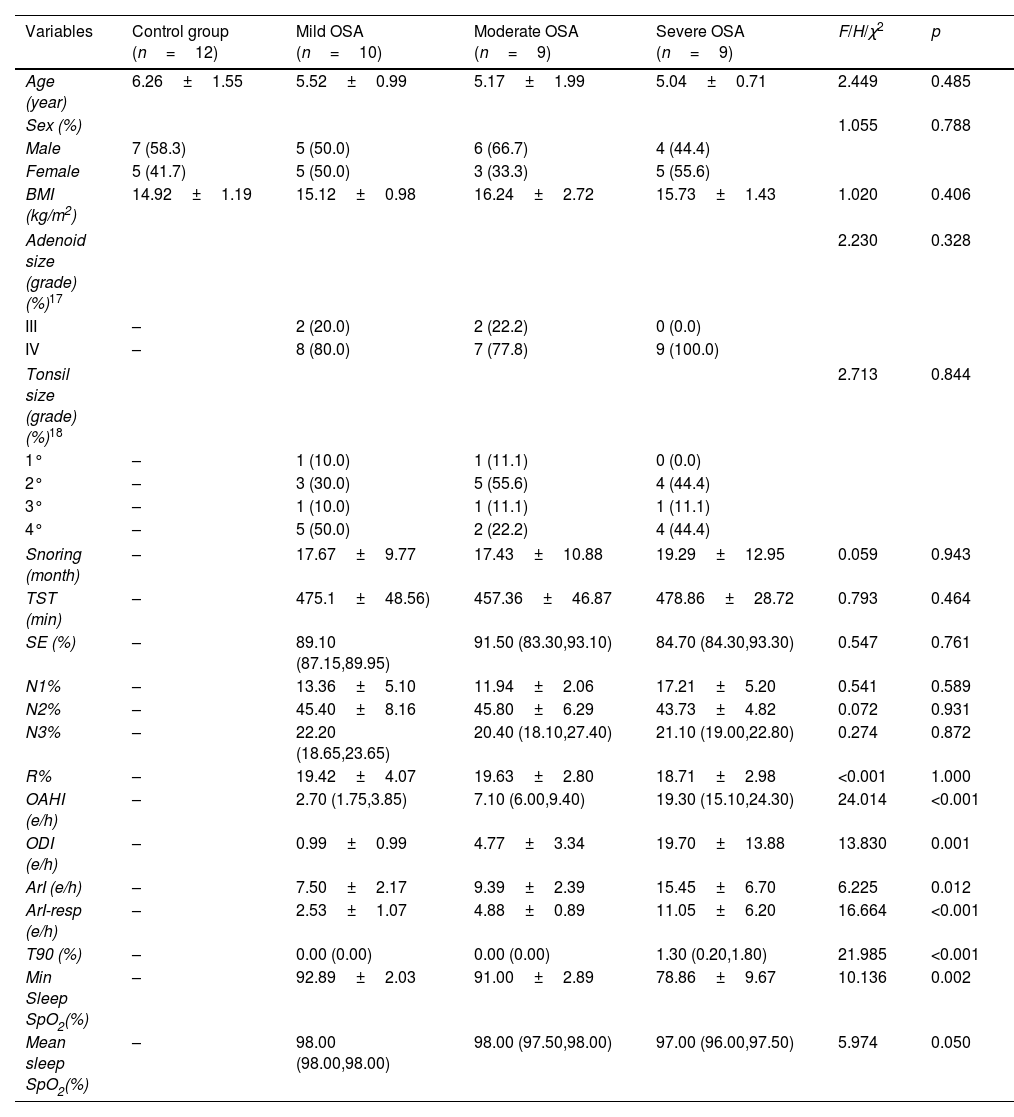
MethodsData-independent acquisition quantitative proteomic analysis was employed to identify serum and urine proteins with differential expression patterns between children with OSA and controls. Differentially expressed proteins that gradually increased or decreased with the severity of OSA were retained as potential biomarkers and underwent ELISA validation.
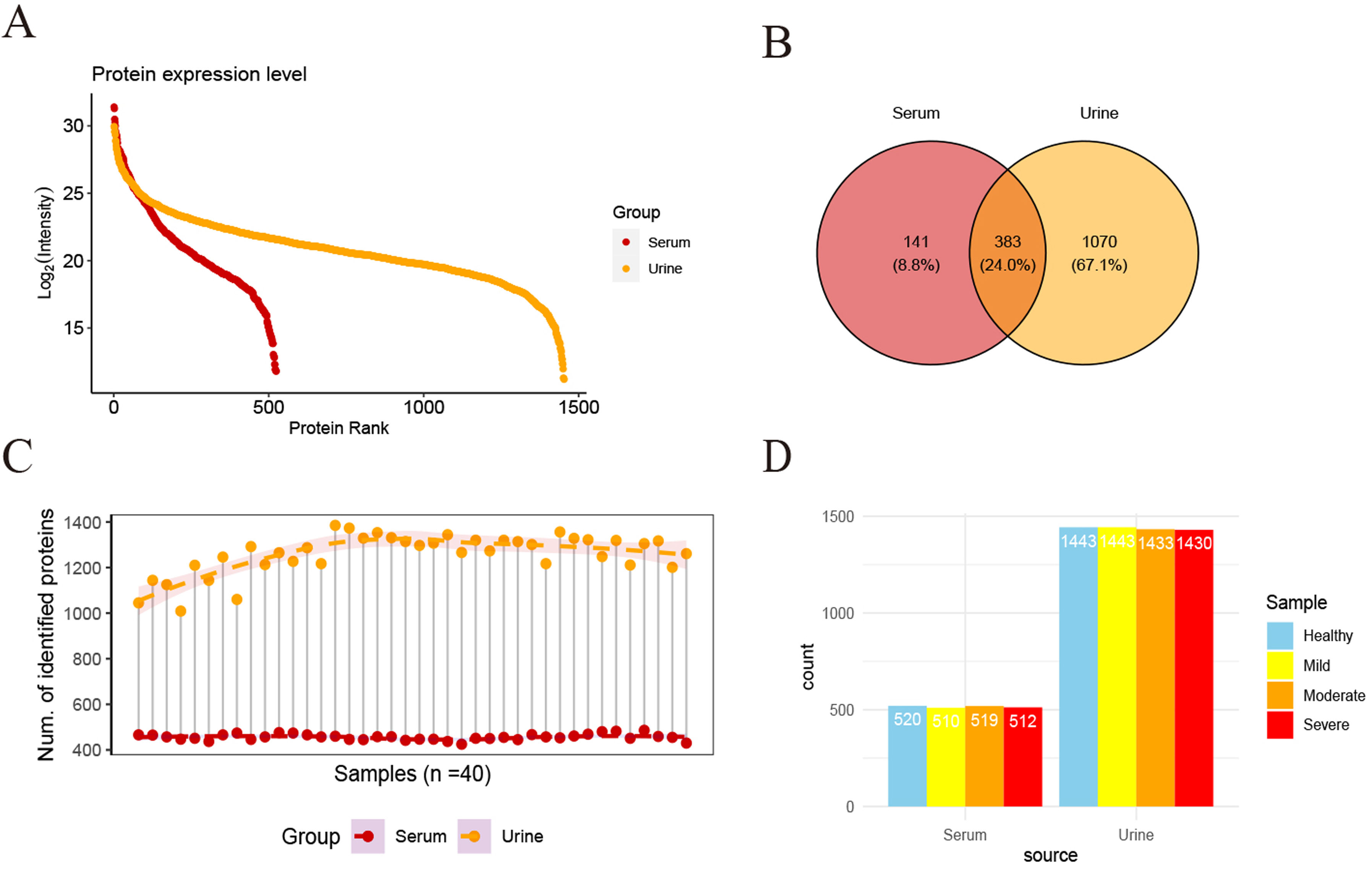
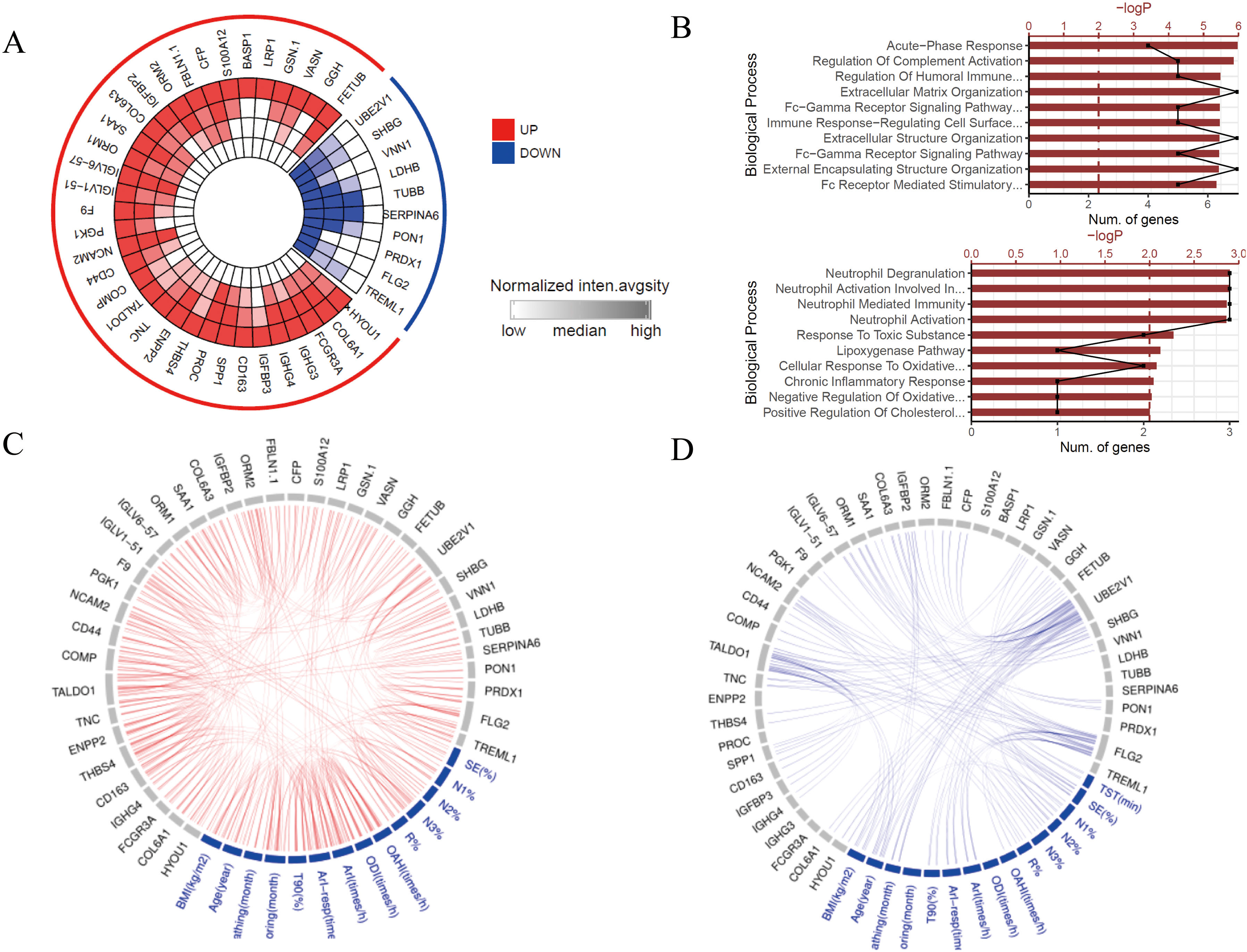
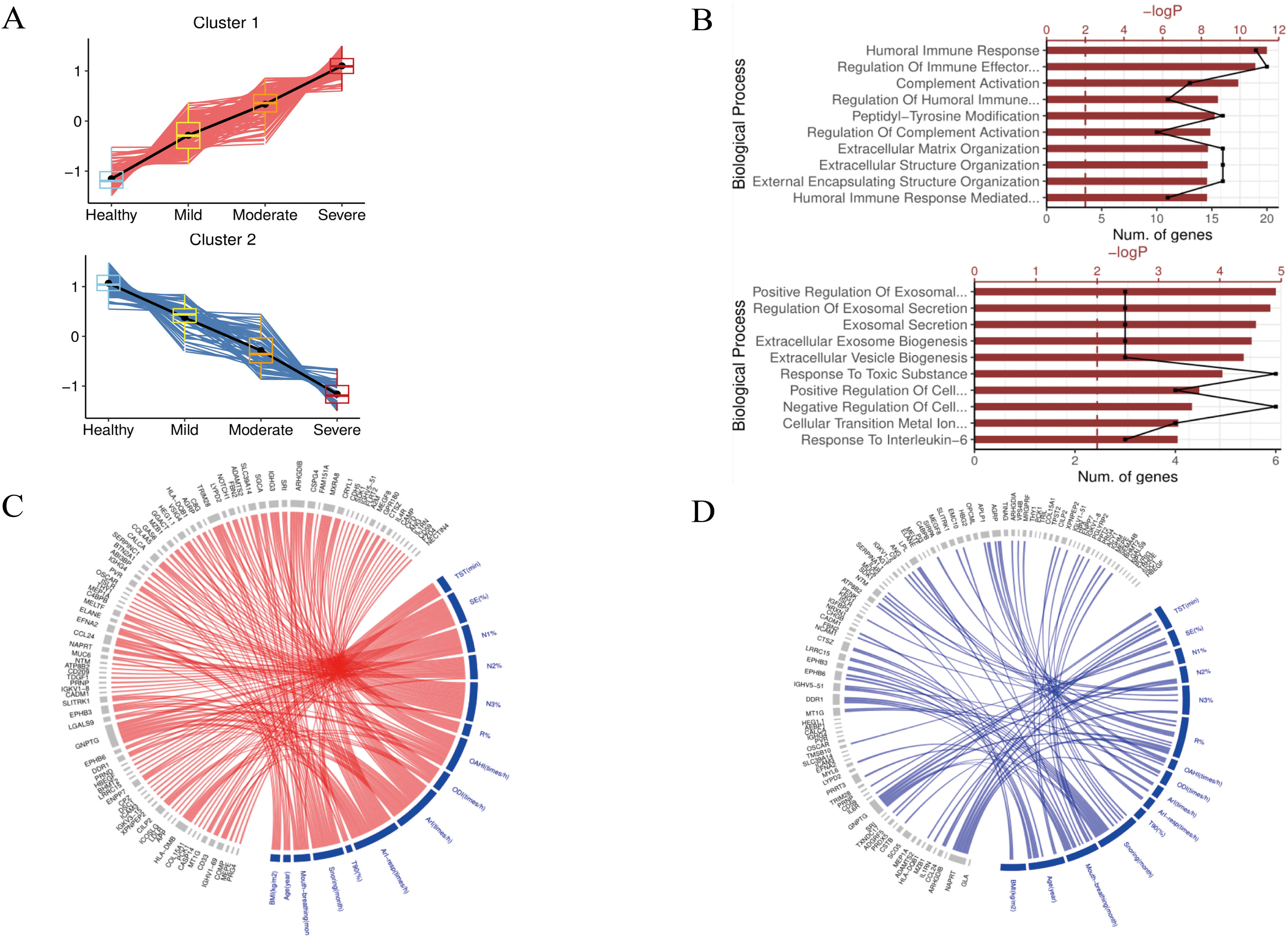
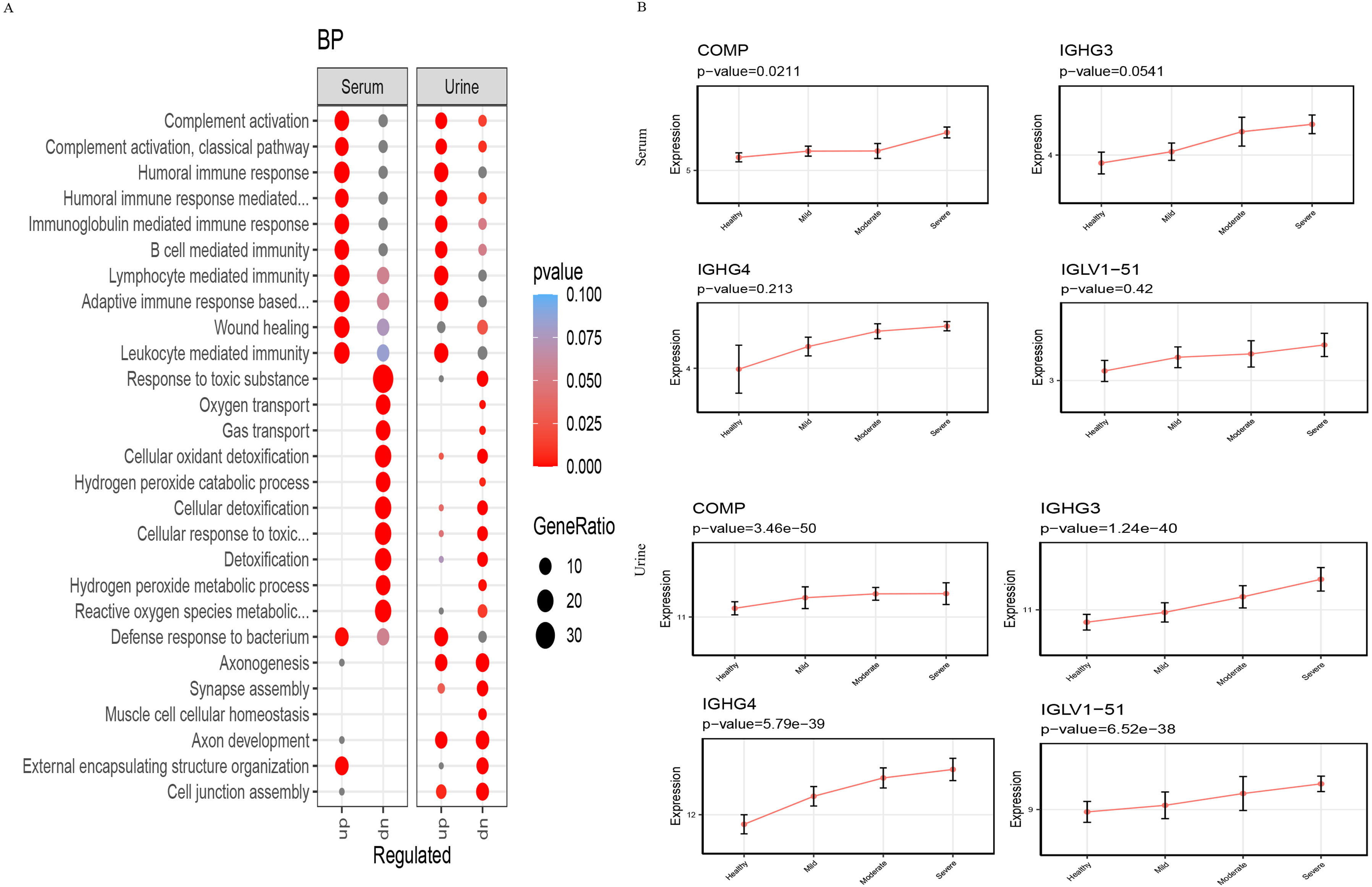
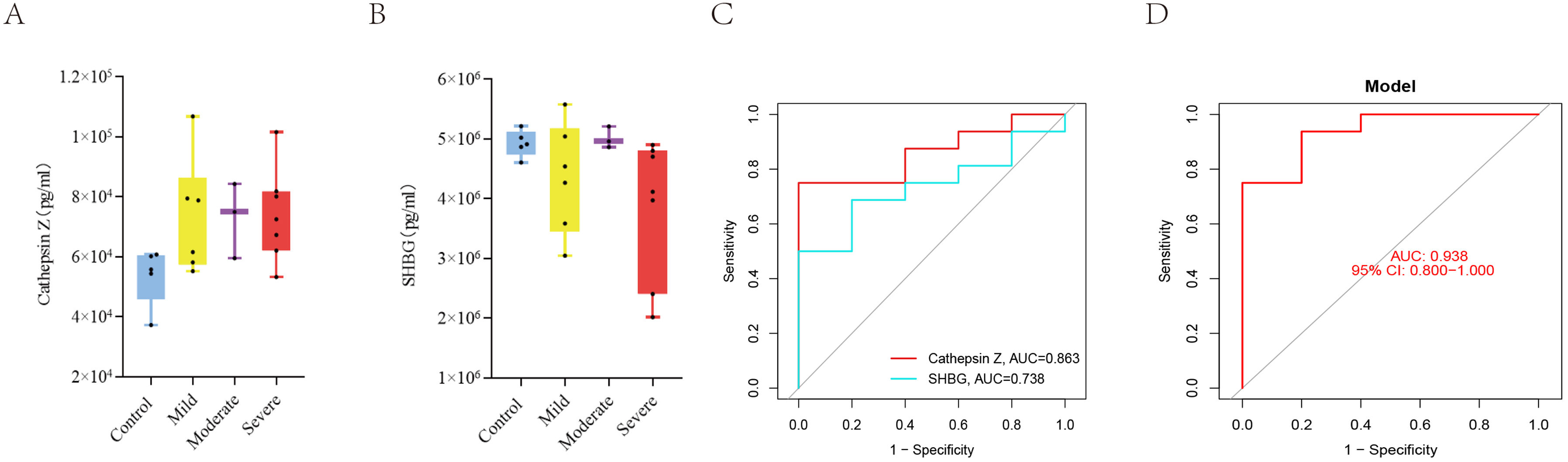
ResultsWe found that with increasing severity of OSA, there was a gradual upregulation of 34 proteins in the serum and 124 proteins in the urine, along with a respective downregulation of 10 serum proteins and 64 urinary proteins in the initial cohort of 40 children. These proteins primarily participate in immune activation, the complement pathway, oxygen transport, and reactive oxygen metabolism. Notably, cathepsin Z exhibited a positive correlation with the obstructive apnea hypopnea index, whereas sex hormone-binding globulin (SHBG) was negatively correlated. These proteins were then validated by ELISA in an independent cohort (n=21). Circulating cathepsin Z and SHBG levels displayed acceptable diagnostic performance of OSA with AUC values of 0.863 and 0.738, respectively.
ConclusionsWe identified two promising circulating proteins as novel biomarkers for clinical diagnosis and assessment of pediatric OSA severity. Furthermore, the comprehensive proteomic profile in pediatric OSA should aid in exploring the underlying pathophysiological mechanisms associated with this prevalent condition.