Asthma and chronic obstructive pulmonary disease (COPD) are common chronic airway diseases that may overlap in some individuals. Asthma COPD overlap (ACO) is a heterogeneous conditions that includes smoking-asthma (SA) and COPD with eosinophilia (COPDe). MicroRNAs (miRNA) are regulators of gene expression with a great potential as biomarkers.
ObjectivesThe objective of this study was to identify distinctive miRNA signatures in patients from the whole spectrum of chronic obstructive bronchial disease (SA, COPDe, non-smoking asthmatics (NSA), and COPD) that could serve as diagnostic biomarkers or describe differential molecular mechanisms with potential therapeutic implications.
MethodsFrom a previously characterized cohort of ACO, COPD and asthma patients, we selected a discovery group of 40 patients for miRNA expression profiling by means of microarray technology. Differential expression of miRNAs were validated by quantitative PCR in the complete cohort (n=274).
ResultsThirty differentially expressed miRNAs (eBAYES p<0.05, fold change ≥2) were found among the different groups of patients regarding COPDe: 19 COPD-vs-COPDe, 13 NSA-vs-COPDe, 11 SA-vs-COPDe. A characteristic down-regulated miRNA expression pattern was identified in COPDe patients. Differential expression of miR-619-5p and miR-4486 in COPDe patients were validated in the complete cohort (n=274).
ConclusionsWe postulate that COPDe patients show a characteristic expression profile of miRNAs distinctive from asthma and COPD. Also that SA and COPDe patients, which have been typically clustered in the ACO group, display distinct molecular events.
El asma y la enfermedad pulmonar obstructiva crónica (EPOC) son enfermedades crónicas comunes de la vía aérea y pueden solaparse en algunos individuos. El solapamiento de asma y EPOC (ACO, por sus siglas en inglés) es una enfermedad heterogénea que incluye el asma en fumadores (AF) y la EPOC con eosinofilia (EPOCe). Los microRNA (miRNA) son reguladores de la expresión de genes con gran potencial para su uso como biomarcadores.
ObjetivosEl objetivo de este estudio fue identificar las firmas características de miRNA en pacientes del espectro de enfermedades pulmonares obstructivas crónicas al completo (AF, EPOCe, asmáticos no fumadores y EPOC) que pudieran servir como biomarcadores diagnósticos o describir mecanismos moleculares diferenciales con potenciales implicaciones terapéuticas.
MétodosA partir de una cohorte previamente caracterizada de pacientes con ACO, EPOC y asma, seleccionamos un grupo de descubrimiento de 40 pacientes para realizar sus perfiles de expresión de miRNA mediante microarrays. La expresión diferencial de miRNA se validó mediante PCR cuantitativa en la cohorte completa (n=274).
ResultadosSe encontraron 30 miRNA expresados diferencialmente (eBayes p<0,05, fold change [cambio en incremento]≥2) entre los diferentes grupos de pacientes en relación con la EPOCe: 19 EPOC comparado con EPOCe, 13 asmáticos no fumadores comparado con EPOCe, 11 AF comparado con EPOCe. Se identificó un patrón característico de expresión con regulación a la baja de miRNA. La expresión diferencial de miR-619-5p y miR-4486 en los pacientes con EPOCe se validó con la cohorte al completo (n=274).
ConclusionesPostulamos que los pacientes con EPOCe muestran un perfil de expresión de miRNA característico y diferente al del asma y la EPOC. También que los pacientes con AF y con EPOCe, que se han agrupado típicamente en el grupo de ACO, muestran eventos moleculares diferenciales.
Asthma and chronic obstructive pulmonary disease (COPD) are the two most common chronic respiratory diseases with different mechanisms and clinical characteristics, although, features of both diseases can be found in some patients. Recently, renewed interest has been raised to the existence of a mixed phenotype called ACO (asthma and COPD overlap).1–3 The specific diagnostic criteria for this overlap are unclear and there is much controversy in the literature upon its existence.4 Moreover, it pools two different conditions, namely smoking asthma (SA) and eosinophilic COPD (COPDe) that are clinically different although little is known about their underlying mechanisms.5 Despite the increasing interest on the analysis of the clinical characteristics of ACO, so far there are few biomarkers able to identify it, probably due to the heterogeneity and biological complexity of this entity.
MicroRNAs (miRNAs) are endogenous single-strand RNA molecules, non-coding, 18–25 nucleotides in length, which regulate post-transcriptional gene expression, interacting through sequence homology with messenger RNAs. Recently, miRNAs have been proposed as molecules with an important role in lung development, as well as in the pathophysiology of various lung diseases.6 MiRNAs have great potential as biomarkers,7 due to their high stability to degradation, circulation in body fluids,8 and easy detection by non-invasive techniques. Differentially expressed miRNAs between healthy and asthmatic and/or COPD patients have already been reported, demonstrating their role in the regulation of different inflammatory processes related to these obstructive pulmonary diseases and airway remodeling.6,9,10 However, the miRNA expression profile for asthma versus COPD was only addressed by Wang et al., identifying seven up/down-regulated miRNAs differentially expressed in the plasma of patients with asthma compared to patients with COPD.11 Additionally, Lacedonia et al. did not observe differences between the expression of miRNAs-145 and miRNA-338 in ACO patients, compared to asthma or COPD patients.12
The main objective of this study is to determine a miRNA molecular signature distinctive of eosinophilic COPD and smoking asthma in order to identify a common molecular mechanism or a potential biomarker that could guide therapy.
Material and methodsStudy designThis cross-sectional, observational, multicenter study was carried out in 23 out-patient clinics from tertiary hospitals in Spain run by expert respiratory physicians. The design of the study has been previously described elsewhere.13 Additionally, an independent Ethics Committee or institutional review board for each study centre approved the final protocol. The STROBE standards for reporting observational studies were followed.
PatientsWe recruited adult patients (≥40 years) who signed an informed, written consent form, diagnosed with chronic bronchial obstructive disease (post-bronchodilator FEV1/FVC≤70%) and divided into 4 groups: asthma (smokers and non-smokers) and COPD (eosinophilic and non-eosinophilic). Patients had to be in a stable condition, free from exacerbations for at least 3 months. Exclusion criteria included primary bronchiectasis, active cancer (metastatic, progressive, or treated within the last 24 months), chronic inflammatory diseases and poor performance status. Active smoking was not an exclusion criterion.
As previously described,13 patients were grouped according to the baseline diagnosis, smoking history and eosinophil counts into four categories: (1) Non-smoking asthmatics (NSA): patients with a history of physician-diagnosed asthma according to international guidelines,14 with chronic airflow obstruction that were either never-smokers or ex-smokers with smoking history of ≤10 pack-years; (2) Smoking asthmatics (SA): asthma patients with chronic airflow obstruction and a smoking history of ≥20 pack-years; (3)COPD was diagnosed according to international recommendations15 by the presence of post-bronchodilator FEV1/FVC ≤0.70 in patients with smoking history of ≥10 pack-years in the absence of a clinical suspicion for asthma or a eosinophil count <200cells/μL in blood; and (4) COPD with eosinophilia (COPDe): COPD patients with eosinophil count ≥200eosinophils/μL in blood. We selected this threshold to recruit the patients because below this cut-off patients are unlikely to have sputum eosinophilia, according to published evidence.16 Recruited patients underwent socio-demographic and clinical questionnaires, lung-function tests and blood extraction. Serum was isolated and frozen at −80°C.
Total RNA isolation from serumTotal RNA including the miRNAs fraction were isolated from 200μL of serum with the miRNeasy serum/plasma kit (Qiagen, Spain) according to the manufacturer's protocol. 108 copies of UniSp6 RNA Spike-in control (Exiqon, Spain) were added to serum before RNA isolation. UniSp6 RNA Spike-in control and the circulating miRNA miR-16-5p were used for monitoring both RNA isolation and cDNA synthesis procedures by quantitative PCR (qPCR).
miRNA expression profiling in serum in a discovery set of patientsA discovery set of samples were selected from patients for microarray analysis. Four groups were considered NSA (n=10), SA (n=10), COPD (n=10) and COPDe (n=10). All groups were matched by sex. NSA and SA groups were also matched by allergy diagnosis (positive or negative skin prick test, n=5 each group). COPD and COPDe groups were matched by currently smokers and non-smokers (n=5 each group).
Accurate miRNA isolation was previously confirmed by qPCR for UniSp6 RNA Spike-in control and for the endogenous miR-16-5p. Cq values were consistent between all 40 samples. Total RNA (8μL) was directly labeled with the FlashTag™ Biotin HSR Labeling Kit and hybridized at 48°C and 60rpm for 42h to the GeneChip® miRNA 4.0 Array (Applied Biosystems, Thermo fisher). Array normalization was executed with the Transcriptome Analysis Software 4.0 (Applied Biosystems, Thermo fisher) following the robust multi-chip analysis (RMA) and the detected above background (DABG) algorithms. The eBayes method was used for identification of differentially expressed miRNAs between groups, considering as biologically significant if displayed a fold change value (FC; <−2 or >2) and p-value <0.05. Venn diagram representation was performed through the Venny.17 Average (Log2) signal value from each group were subjected to unsupervised hierarchical clustering using average linkage and Euclidian distance as a similarity metric to visualize likenesses among miRNA and pathologies (MultiExperiment Viewer MeV 4.9.0 software). Clustering results were combined in a two-dimensional heat map with color intensities according to the pattern of miRNA expression. DIANA-miRPath v3.0 were used for KEGG pathway enrichment based in predicted miRNA targets provided by the DIANA-microT-CDS algorithm.18
Real time quantitative PCRWe used the miRCURY LNA™ Universal RT microRNA PCR System (Exiqon, Qiagen, Spain) for the detection of miRNAs by quantitative real-time PCR (qPCR) using SYBR green. Total RNA (2μL) obtained from serum were transcribed to cDNA with the universal cDNA synthesis kit, diluted 20× with nuclease free water and real-time PCR amplification with LNA™ specific primers using a CFX96 system (Bio-Rad, Sapin). Product specificity was confirmed in initial experiments by melting curve analysis and PCR efficiency was calculated for each LNA™ specific primer (100±5%). Relative expression of the miRNA of interest was calculated using the 2−ΔΔCq method.19 MiRNAs Cq values were normalized respected to miR-16-5p Cq values.
Statistical analysisData are represented as mean±standard error of mean (SEM). For multiple comparisons between groups Kruskal–Wallis test was used and Mann–Whitney U-test was used for compare differences between two independent groups. The significance level was established as a two-tailed p-value <0.05. miRNAs performance was evaluated by receiver operating curve (ROC) analysis. For better discrimination of COPDe group versus the rest of pathologies, principal component analysis was performed. Also a multivariate logistic regression analysis was created using the cut-off values of miR-619-5p and miR-4486 expression defined by the Youden index. Age, sex, blood eosinophil count and percentage of post-bronchodilator FEV1 were included as confounding variables. Statistical analyses were performed with the SPSS Statistic 17.0 Software.
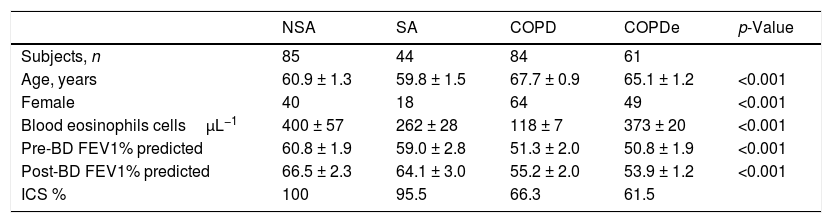
ResultsPatient characteristics by groupTwo hundred and seventy-four patients with asthma, COPD and ACO were included in this study (85 NSA, 44 SA, 84 COPD and 61 COPDe). Extensive characterization of the demographic, clinical and functional features of the entire population were reported in.13,20 A summary of the most prominent clinical charateristics are shown in Table 1. Briefly, the degree of bronchial obstruction was moderate and symptoms were fairly controlled as assessed by the CAT and ACT questionnaires (data not shown). Most patients (80%) were treated with ICS (distribution by groups in Table 1) and almost all of them received a long-acting β2-agonist (LABA). Six patients in the NSA group were receiving oral corticosteroids. Patients classified as COPDe showed significantly lower post-bronchodilator FEV1 than SA patients (53.94±2.1% vs. 64.1±3.06%, Mann–Whitney test; p<0.005). Albeit COPDe patients were previously sorted as blood eosinophil count ≥200cells/μL, this group presented higher levels for blood eosinophils count than SA patient group (373±20 vs. 262±28cells/μL, Mann–Whitney test; p<0.001).
Characteristics of patients according to the baseline diagnosis.
| NSA | SA | COPD | COPDe | p-Value | |
|---|---|---|---|---|---|
| Subjects, n | 85 | 44 | 84 | 61 | |
| Age, years | 60.9 ± 1.3 | 59.8 ± 1.5 | 67.7 ± 0.9 | 65.1 ± 1.2 | <0.001 |
| Female | 40 | 18 | 64 | 49 | <0.001 |
| Blood eosinophils cellsμL−1 | 400 ± 57 | 262 ± 28 | 118 ± 7 | 373 ± 20 | <0.001 |
| Pre-BD FEV1% predicted | 60.8 ± 1.9 | 59.0 ± 2.8 | 51.3 ± 2.0 | 50.8 ± 1.9 | <0.001 |
| Post-BD FEV1% predicted | 66.5 ± 2.3 | 64.1 ± 3.0 | 55.2 ± 2.0 | 53.9 ± 1.2 | <0.001 |
| ICS % | 100 | 95.5 | 66.3 | 61.5 |
Data are presented as mean±SD or percentages, unless otherwise stated. NSA: non-smoking asthmatics, SA: smoking asthmatics; COPD: chronic obstructive pulmonary disease; COPDe: eosinophilic COPD; pre-BD: pre-bronchodilator; FEV1: forced expiratory volume in 1s; post-BD: post-bronchodilator; ICS: inhaled corticosteroids. p-value represents t-test for differences between means and Chi-squared test for differences between proportions.
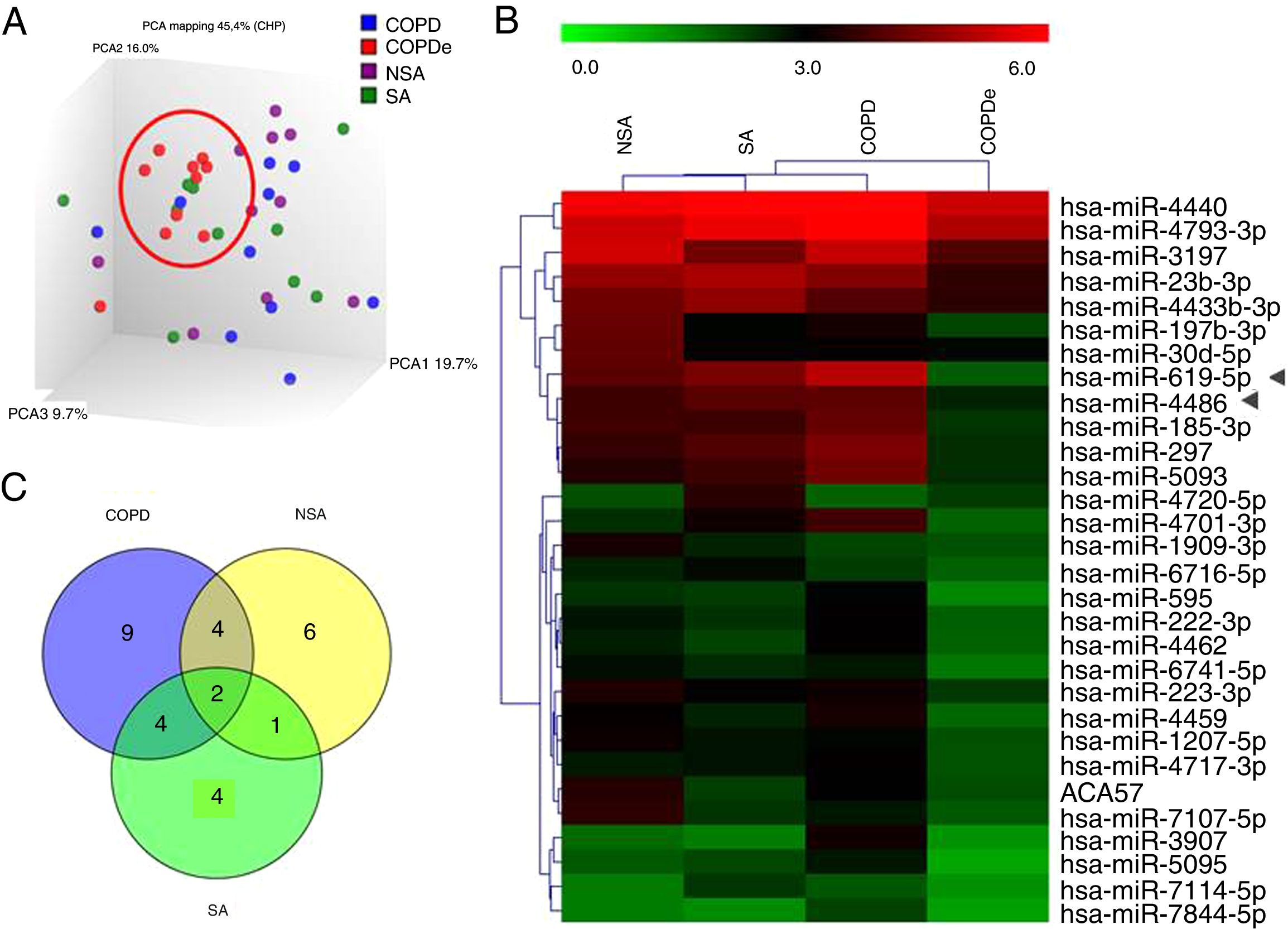
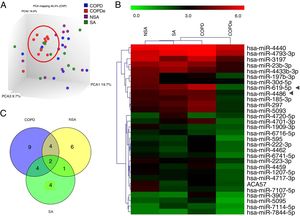
A discovery set of samples was selected from patients for microarray analysis considering the four groups: NSA, SA, COPD and COPDe. Principal component analysis of the normalized signal values from the 40 samples showed that COPDe samples appeared to be more consistently grouped. Such pattern could not be observed in the remaining pathologies (Fig. 1A).
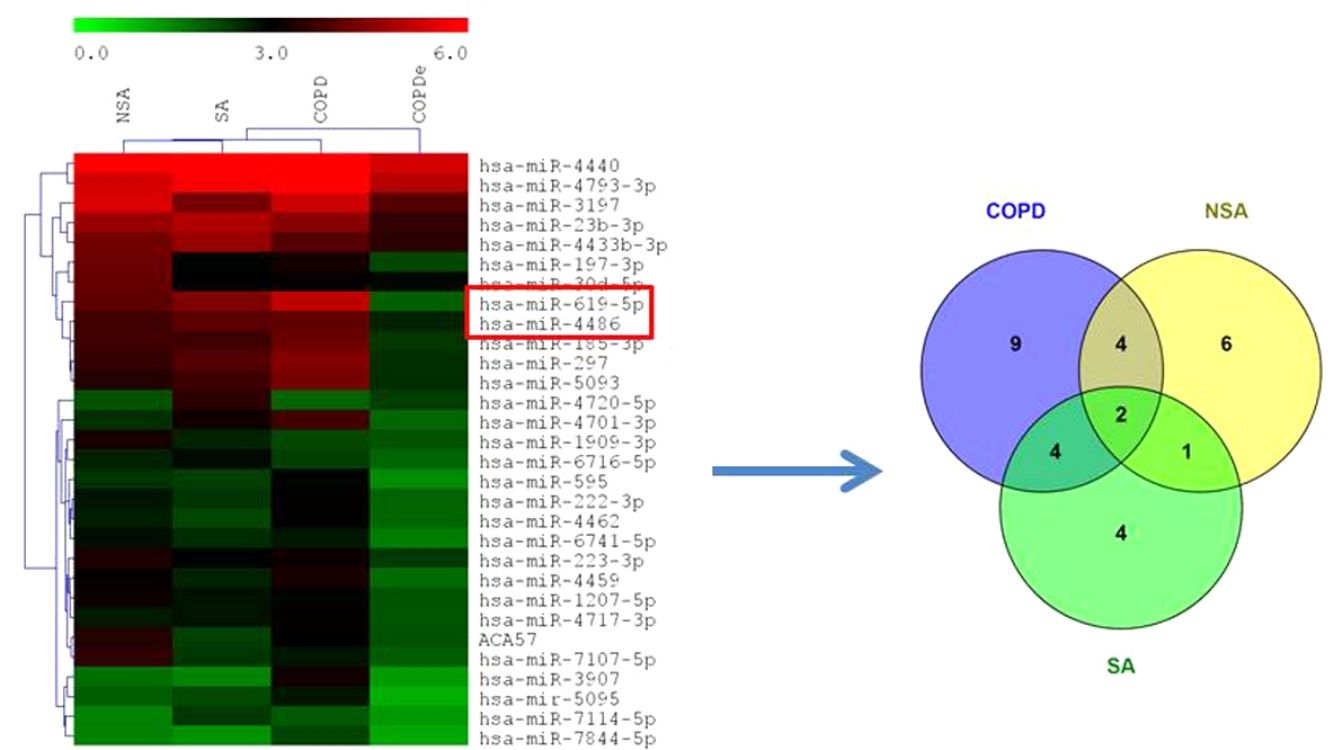
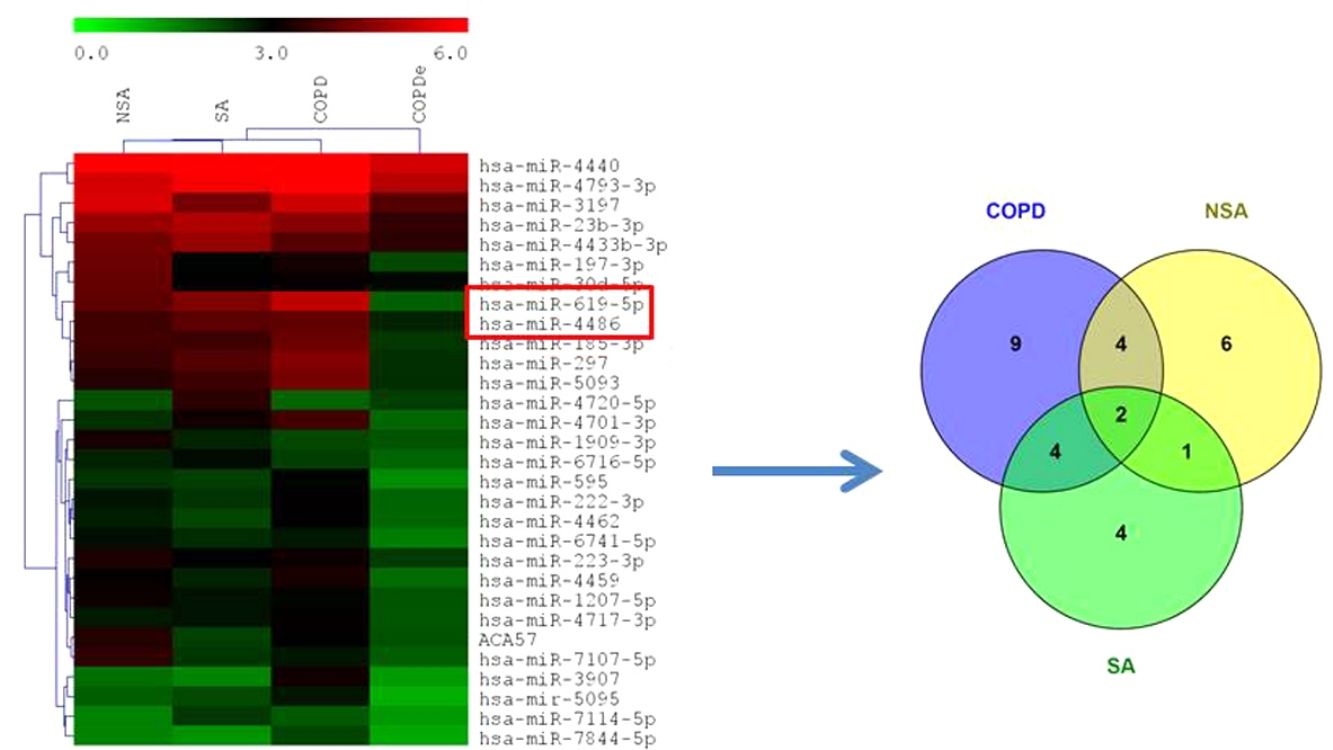
MiRNA microarray profiling of NSA, SA, COPD and COPDe discovery groups. (A) Three-dimensional principal component analysis (PCA) plot showing the normalized signal values of the 40 microarray from non-smoking asthmatics (NSA), smoking asthmatic (SA), chronic obstructive pulmonary disease (COPD) and eosinophilic COPD (COPDe) patients. (B) Heat map representation of an unsupervised hierarchical clustering of the 30 significant differentially expressed microRNAs in comparisons including COPDe group. Arrow heads show miR-619-5p and miR-4486 miRNAs. (C) Venn diagram of significant differentially expressed miRNAs in all pairwise comparisons including COPDe.
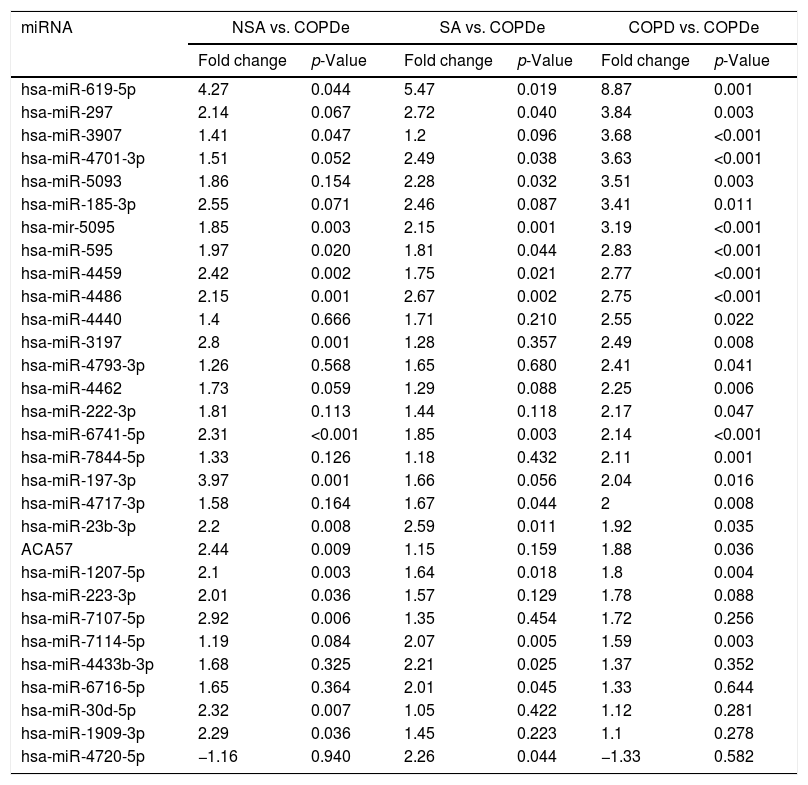
Significant differences in miRNA expression were mostly found in comparison analysis between the COPDe group and the remaining pathologies. Overall, 29 mature miRNAs and one small nucleolar RNA were down-regulated in the COPDe group: 19 miRNAs in comparison with COPD, 13 with NSA, and 11 with SA, respectively (see Table 2). A heatmap of unsupervised clustering analysis of all significant differentially expressed miRNAs related to COPDe is shown in Fig. 1B. However, only miR-619-5p and miR-4486 were consistently significant in all the comparisons between the COPDe group and the remaining pathologies, as shown in the Venn diagram in Fig. 1C.
Differentially expressed miRNAs in comparisons including COPDe.
| miRNA | NSA vs. COPDe | SA vs. COPDe | COPD vs. COPDe | |||
|---|---|---|---|---|---|---|
| Fold change | p-Value | Fold change | p-Value | Fold change | p-Value | |
| hsa-miR-619-5p | 4.27 | 0.044 | 5.47 | 0.019 | 8.87 | 0.001 |
| hsa-miR-297 | 2.14 | 0.067 | 2.72 | 0.040 | 3.84 | 0.003 |
| hsa-miR-3907 | 1.41 | 0.047 | 1.2 | 0.096 | 3.68 | <0.001 |
| hsa-miR-4701-3p | 1.51 | 0.052 | 2.49 | 0.038 | 3.63 | <0.001 |
| hsa-miR-5093 | 1.86 | 0.154 | 2.28 | 0.032 | 3.51 | 0.003 |
| hsa-miR-185-3p | 2.55 | 0.071 | 2.46 | 0.087 | 3.41 | 0.011 |
| hsa-mir-5095 | 1.85 | 0.003 | 2.15 | 0.001 | 3.19 | <0.001 |
| hsa-miR-595 | 1.97 | 0.020 | 1.81 | 0.044 | 2.83 | <0.001 |
| hsa-miR-4459 | 2.42 | 0.002 | 1.75 | 0.021 | 2.77 | <0.001 |
| hsa-miR-4486 | 2.15 | 0.001 | 2.67 | 0.002 | 2.75 | <0.001 |
| hsa-miR-4440 | 1.4 | 0.666 | 1.71 | 0.210 | 2.55 | 0.022 |
| hsa-miR-3197 | 2.8 | 0.001 | 1.28 | 0.357 | 2.49 | 0.008 |
| hsa-miR-4793-3p | 1.26 | 0.568 | 1.65 | 0.680 | 2.41 | 0.041 |
| hsa-miR-4462 | 1.73 | 0.059 | 1.29 | 0.088 | 2.25 | 0.006 |
| hsa-miR-222-3p | 1.81 | 0.113 | 1.44 | 0.118 | 2.17 | 0.047 |
| hsa-miR-6741-5p | 2.31 | <0.001 | 1.85 | 0.003 | 2.14 | <0.001 |
| hsa-miR-7844-5p | 1.33 | 0.126 | 1.18 | 0.432 | 2.11 | 0.001 |
| hsa-miR-197-3p | 3.97 | 0.001 | 1.66 | 0.056 | 2.04 | 0.016 |
| hsa-miR-4717-3p | 1.58 | 0.164 | 1.67 | 0.044 | 2 | 0.008 |
| hsa-miR-23b-3p | 2.2 | 0.008 | 2.59 | 0.011 | 1.92 | 0.035 |
| ACA57 | 2.44 | 0.009 | 1.15 | 0.159 | 1.88 | 0.036 |
| hsa-miR-1207-5p | 2.1 | 0.003 | 1.64 | 0.018 | 1.8 | 0.004 |
| hsa-miR-223-3p | 2.01 | 0.036 | 1.57 | 0.129 | 1.78 | 0.088 |
| hsa-miR-7107-5p | 2.92 | 0.006 | 1.35 | 0.454 | 1.72 | 0.256 |
| hsa-miR-7114-5p | 1.19 | 0.084 | 2.07 | 0.005 | 1.59 | 0.003 |
| hsa-miR-4433b-3p | 1.68 | 0.325 | 2.21 | 0.025 | 1.37 | 0.352 |
| hsa-miR-6716-5p | 1.65 | 0.364 | 2.01 | 0.045 | 1.33 | 0.644 |
| hsa-miR-30d-5p | 2.32 | 0.007 | 1.05 | 0.422 | 1.12 | 0.281 |
| hsa-miR-1909-3p | 2.29 | 0.036 | 1.45 | 0.223 | 1.1 | 0.278 |
| hsa-miR-4720-5p | −1.16 | 0.940 | 2.26 | 0.044 | −1.33 | 0.582 |
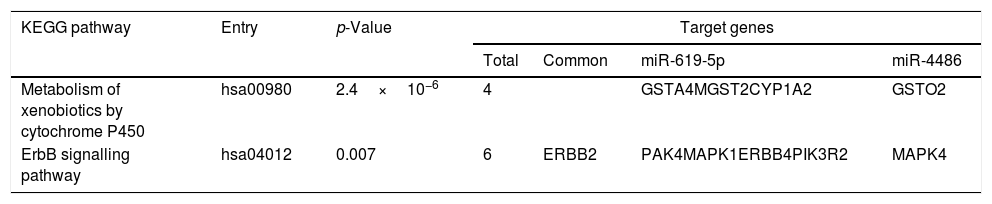
Predicted target genes of miR-619-5p and miR-4486 were identified with DIANA tools. Pathway union enrichment analysis revealed that both miRNAs participate in two KEGG pathways: “Metabolism of xenobiotics by cytochrome P450” and “ErbB signalling pathway” by targeting four and six genes related to these pathways, respectively. Erbb2 is a common predicted target gene of both miR-619-5p and miR-4486 (Table 3).
Functional analysis of common predicted gene targets for miR-619-5p and miR-4486.
| KEGG pathway | Entry | p-Value | Target genes | |||
|---|---|---|---|---|---|---|
| Total | Common | miR-619-5p | miR-4486 | |||
| Metabolism of xenobiotics by cytochrome P450 | hsa00980 | 2.4×10−6 | 4 | GSTA4MGST2CYP1A2 | GSTO2 | |
| ErbB signalling pathway | hsa04012 | 0.007 | 6 | ERBB2 | PAK4MAPK1ERBB4PIK3R2 | MAPK4 |
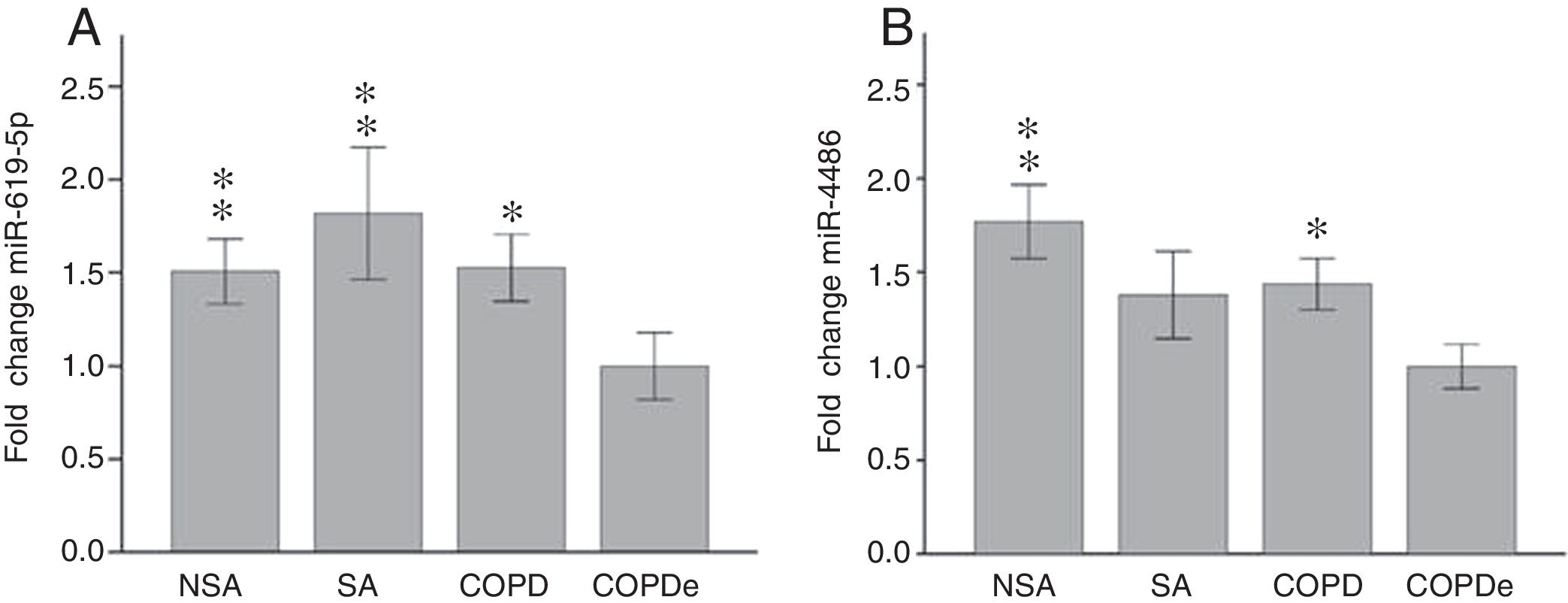
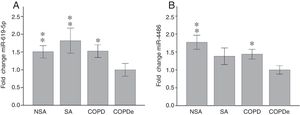
Once identified miR-619-5p and miR-4486 as possible candidates able to distinguish COPDe patients from the rest of patients by microarray analysis (40 samples), we proceeded to validate them in the complete cohort of 274 patients by qPCR. Expression levels of miR-619-5p and miR-4486 were measured in 85 NSA, 44 SA, 84 COPD and 61 COPDe serum samples. MiR-619-5p was significantly down-regulated in COPDe group compared with the rest of the pathologies (Fig. 2A), while miR-4486 was significantly down-regulated in COPDe patients compared with NSA and COPD patients (Fig. 2B).
MiR-619-5p and miR-4486 expression in patients groups of the complete CHACOS cohort (n=274). NSA: non-smoking asthmatics, SA: smoking asthmatics; COPD: chronic obstructive pulmonary disease; COPDe: eosinophilic COPD. Results are represented as means±SEM. *p<0.05, **p<0.01, when compared to COPDe group.
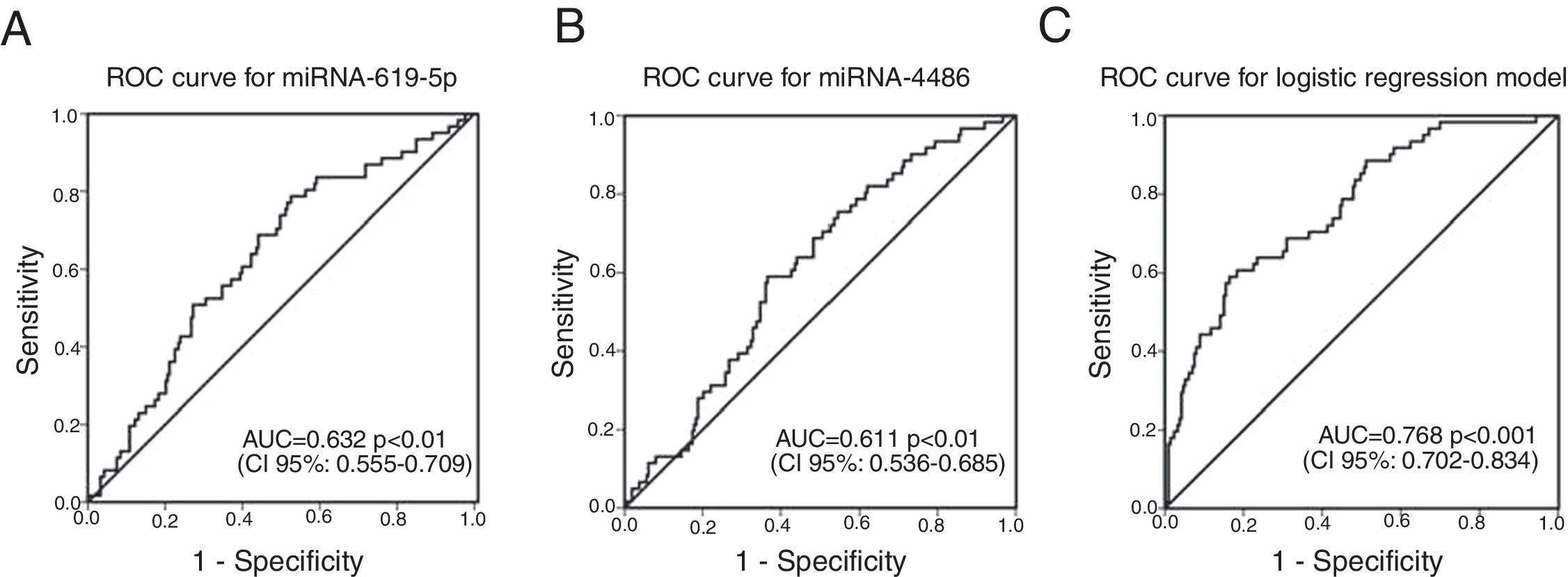
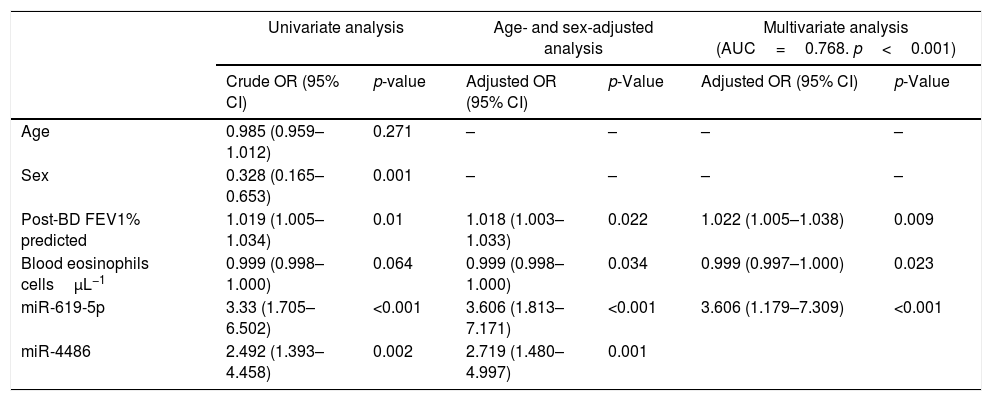
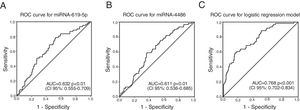
ROC curves were determined for COPDe discrimination. Overall, miR-619-5p and miR-4486 significantly discriminate (p<0.001) COPDe patients from the rest of patients with AUCs of 0.63 and 0.61, respectively (Fig. 3A and B). Multivariate logistic regression analysis of risk factors associated to COPDe was performed. Age, sex, blood eosinophil count, percentage of post-bronchodilator FEV1, and miR-619-5p were included in the model. MiR-4486 was not significant and, therefore, excluded from the model. Crude and adjusted Odds Ratio (O.R.) are indicated in Table 4. Comparison of the expected and observed frequencies by the Hosmer–Lemeshow goodness-of-fit test (p-value <0.001) and by ROC curve (AUC=0.768; p<0.001) indicated a good fit for the model (Fig. 3C).
Multivariate logistic regression of risk factors associated to COPDe.
| Univariate analysis | Age- and sex-adjusted analysis | Multivariate analysis (AUC=0.768. p<0.001) | ||||
|---|---|---|---|---|---|---|
| Crude OR (95% CI) | p-value | Adjusted OR (95% CI) | p-Value | Adjusted OR (95% CI) | p-Value | |
| Age | 0.985 (0.959–1.012) | 0.271 | – | – | – | – |
| Sex | 0.328 (0.165–0.653) | 0.001 | – | – | – | – |
| Post-BD FEV1% predicted | 1.019 (1.005–1.034) | 0.01 | 1.018 (1.003–1.033) | 0.022 | 1.022 (1.005–1.038) | 0.009 |
| Blood eosinophils cellsμL−1 | 0.999 (0.998–1.000) | 0.064 | 0.999 (0.998–1.000) | 0.034 | 0.999 (0.997–1.000) | 0.023 |
| miR-619-5p | 3.33 (1.705–6.502) | <0.001 | 3.606 (1.813–7.171) | <0.001 | 3.606 (1.179–7.309) | <0.001 |
| miR-4486 | 2.492 (1.393–4.458) | 0.002 | 2.719 (1.480–4.997) | 0.001 | ||
Crude and adjusted odds ratio (OR) are indicated in the table. Comparison of the expected and observed frequencies by the Hosmer–Lemeshow goodness-of-fit test and by ROC curve indicated a good fit for the model. AUC: area under the curve; post-BD: post-bronchodilator; FEV1: forced expiratory volume in 1s.
The main result of this work is the characterization, for the first time, of a specific miRNA profile differentially expressed in patients with eosinophilic COPD that is different to that shown by smoking asthmatics, NSA or COPD.
Although the diagnosis of ACO by clinical criteria includes both, smoking asthmatics (SA) and COPD patients with eosinophilia (COPDe), for better discrimination purposes, we have considered SA and COPDe as two different entities based on previous work from our group showing that SA and COPDe patients displayed significant differences in demographic and respiratory functional characteristics.13 Therefore, suggesting that different molecular events may underlie both pathological conditions. These differences between COPDe and SA has also been recently reported from the clinical point of view.21
Overall, 30 miRNAs appeared down-regulated in serum of COPDe patients in comparison with other pathologies (19 COPD, 13 non-smoking asthma, and 11 smoking asthma). Global down-regulation of miRNAs has been described in lung tissue from individuals with COPD compared to those with normal lung.22
Several studies have reported the existence of differentially expressed miRNAs among individuals affected by asthma or COPD in comparison with healthy individuals finding a variable number of miRNAs associated, depending on the type of sample analyzed.6,9 Misregulation of miRNAs expression was seen in a number of different tissues and inflammatory cell types in lung, and these changes correlated with disease severity/risk in a number of studies.10,23 To our knowledge, the comparison of miRNA levels in serum of asthmatic versus COPD patients has only been addressed by Wang et al.,11 who found 18 and 57 differentially expressed miRNAs, respectively. A subset of the most significant differentially expressed miRNAs was determined in a small cohort including ACO patients; however, no significant differences in expression of these miRNAs could be found between ACO and asthma or COPD.12 In a larger cohort of patients, we have been able to identify significant differences between ACO patients. Moreover, we have confirmed the molecular heterogeneity of ACO pathology and differentiated its entities (SA and COPDe). In COPDe, miR-619-5p was consistently found down-regulated compared to COPD, NSA, and SA. For miR-4486, only comparisons between COPDe and NSA or COPD groups were significantly different; however, such differences could not be statistically confirmed between COPDe and SA groups. Altogether, these results suggest that the physiopathology of COPDe has a distinctive molecular signature at the miRNA expression level.
Tobacco exposure could be the triggering factor for the miRNA down-regulation.6,23 In fact, several authors have shown that tobacco smoke causes changes in gene expression of respiratory epithelium, which is linked to the development of respiratory diseases such as COPD.22,24–26 Most of these miRNAs were down-regulated in smokers and inversely correlated with gene expression levels. Our results, however, cannot be explained by a direct relation between miRNA down-regulation and tobacco exposure observed in COPDe patients, because of COPD and smoking asthma patients, also had a current or former history of smoking. Therefore, other molecular conditions specific of the COPDe pathology group must be considered.
Functional analysis of common predicted gene targets for miR-619-5p and miR-4486 revealed that both miRNAs could participate in the regulation of the “ErbB signaling pathway” and “Metabolism of xenobiotics by cytochrome P450” by targeting 6 (ERBB2 and ERBB4, MAPK1 and MAPK4, PAK4 and PIK3R2) and 4 (GSTA4, MGST2, CYP1A2 and GSTO2) genes involved in these pathways, respectively. Epidermal growth factor (EGF) receptors include HER1 (EGFR/ErbB1), HER2 (ErbB2), HER3 (ErbB3) and HER4 (ErbB4). Dysregulation of the EGFR pathway causes aberrant EGFR signaling which is associated with the early stage pathogenesis of respiratory diseases such as lung fibrosis, cancer and numerous airway hypersecretory diseases, including COPD, asthma and cystic fibrosis.27–30 “Metabolism of xenobiotics by P450” appeared as one of the top-ranked COPD-related metabolic networks using disease related genes and their direct interactors in an integrated human metabolic network obtained from merging multiple databases.31 Biotransformation of xenobiotics leads to the generation of reactive oxygen species (ROS). In COPD patients, the protective antioxidant levels are significantly depleted in alveolar macrophages.32 One of the key regulators of this endogenous antioxidant system is the transcription factor NFE2L2/NRF2 (nuclear factor, erythroid-derived 2-like 2). There is significant data suggesting a critical role for NRF2 in preventing lung disease. Studies in COPD patients have shown that NRF2 dependent genes are activated in disease.32 Although accurate functional studies should be performed to validate this in silico results, we suggest that targeting the EGFR signalling pathway or NRF2 modulators could be used as novel therapeutic approaches for treating these respiratory diseases.
MiR-619-5p and miR-4486 were evaluated as potential serum biomarkers to distinguish COPDe from asthmatic and COPD subjects; however, low discriminatory power was obtained for each miRNA individually. In contrast, an integrated logistic regression model showed an adequate discriminatory potential of miR-619-5p to distinguish eosinophilic COPD patients from asthmatic and COPD patients and, particularly from smoking asthmatic patients.
Our study has several limitations. First, there is no gold standard for the diagnosis of asthma in a COPD population. Second, the cut-off eosinophil point to define COPDe is arbitrary; a threshold of 200eosinophils/μL was chosen following previous studies.13 Also, the smoking exposure of 20 packs-year in the asthma population could lead to a selection bias. Third, our patients were recruited from public health services with universal health care and they were receiving treatment for COPD or asthma according to clinical practice; this could affect the results of the clinical outcomes like blood eosinophils or exacerbations.
ConclusionsTo our knowledge, this is the first study to show significant differences in expression at the miRNA level between COPDe and asthmatic or COPD patients. In addition, our results have shown that SA and COPDe patients, who have been typically clustered in the ACO group because they share a similar therapeutic approach, display distinct molecular events. Thus, suggesting that different pathophysiological mechanisms may underlie these respiratory diseases and therefore, different diagnosis and treatment approaches should be considered for smoking asthma and COPD with eosinophilia patients.
AuthorshipVJA, AT, and BGC designed the study, analyzed the results and wrote the manuscript. BGC and LPLL designed the CHACOS study. VJA, AT, and AI performed the technical methodologies. BGC, LPLL and VP recruited patients and reviewed the manuscript. The rest of the CHACOS study team recruited patients.
FundingThe project was endorsed by the COPD and Asthma Research Board of the Spanish Society of Respiratory Medicine (Separ). The project was partially funded by the Fondo de Investigaciones Sanitarias, Instituto de Salud Carlos III, Ministerio de Economia y Competitividad (PI15/01263) through European Union FEDER funds, by a grant from CIBER de Enfermedades Respiratorias, CIBERES and by an unrestricted grant from Chiesi España S.A.U. None of the funding body had any role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of interestVA, AI, AT declare no competing interests. VdP declares personal fees from Astrazeneca outside the submitted work. Dr. Pérez de Llano reports grants from Separ (Spanish Society of Respiratory Medicine and Thoracic Surgery) during the conduct of the study; grants and personal fees from Novartis, personal fees from Chiesi, personal fees from Astrazeneca, personal fees from Pfizer, outside the submitted work. Dr. Cosio reports grants from SEPAR (Sociedad Española de Neumología y Cirugía torácica) during the conduct of the study; personal fees from AstraZeneca, grants from Boehringer, grants and personal fees from Novartis, grants and personal fees from Chiesi, personal fees from Rovi, grants from Menarini, personal fees from Esteve, outside the submitted work.
Authors are grateful to all the patients that participated in the study, and to Dr. Pilar Sanchís from IdISBa (Mallorca, Spain) for her statistical advice.
DUE Rocio Cordova, Hospital Universitario Son Espases-IdISBa, CIBERES, Palma de Mallorca, Spain; Dr. Sergi Pascual, Dra. Mireia Admetlló, Hospital del Mar, Barcelona, Spain; Dra. Salud Santos, Dr. Gianluca Cotta Ramusino, Hospital Universitari de Bellvitge, Barcelona, Spain; Dra. Carmen Gómez Neira, Dra. Irene Carla Martin Robles, Hospital Universitario Lucus Augusti, Lugo, Spain; Dr. Alberto Fernández-Villar, Dra. Cristina Represas, Dra. Ana Priegue Carreracon Hospital Xeneral de Vigo, Vigo, Spain; Dr. Antolin Lopez Viña, Dra. Andrea Trisán Alonso Hospital Puerta del Hierro, Madrid, Spain; Dr. Francisco García Río, Dr. Raúl Galera Martínez, Dra. Raquel Casitas Mateo, Dra. Elisabet Martínez Cerón, Hospital La Paz, Madrid, Spain; Dra. Sagrario Mayoralas, Dr. Adalberto Pacheco Galván, Hospital Universitario Ramón y Cajal, Madrid, Spain; Dr. Luis Puente Maestu, Dra. Julia Garcia de Pedro, Dra. Pilar Sanz, Hospital General Universitario Gregorio Marañón, Madrid, Spain; Dr. Juan Jose Soler-Cataluña, Dr. José Belda Ramírez, Dr. Fernando Sánchez-Toril López, Dr. Rafael Peris Cardells Hospital Arnau de Vilanova, Valencia, Spain; Dra. Eva Martínez Moragón, Dra. Maria Climent Gregori, Hospital Universitario Dr. Peset, Valencia, Spain; Dra. Patricia Garcia-Sidro, Hospital Castellón de la Plana; Dra. Cleofe Fernández, Dra. Mª Encarnación Barroso Medel, Hospital General Universitario de Alicante, Alicante, Spain; Dr. Jose Luis López-Campos, Dra. Carmen Calero, Dra. María Abad Arranz, Dr. Eduardo Márquez Martín, Dr. Francisco Ortega Ruiz Hospital Virgen del Rocío, Sevilla, Spain; Dr. Bernardino Alcazar, Hospital Alta Resolución de la Loja, Granada, Spain; Dr. Javier Callejas, Raúl Godoy Mayoral, Javier Cruz Ruiz, Hospital General Universitario de Albacete, Albacete, Spain; Dra. Nuria Marina Malanda, Dr. Juan Bautista Gáldiz Iturri, Dra. Elena López de Santa Maria Miró, Hospital Universitario Cruces, Bilbao, Spain; Dr. Juan Luis García Rivero, Dra. Ruth Valerie Alain Mbessekg, Hospital de Laredo, Laredo, Spain; Dra. Cristina Martinez, Dra. Rosirys Mercedes Guzmán Taveras, Dra. Ana Fernandez Tena, Dra. Rosario Madiedo de la Llera, Dra. Ángeles Encarnación Malmierca de Dios Instituto Nacional de Silicosis, Oviedo, Spain; Dr. Jose Antonio Ros Lucas, Dra. M. Carmen Soto Fernández, Dr. Rubén Andújar Espinosa, Hospital Universitario Virgen de la Arrixaca, Murcia, Spain; Dr. Marc Miravitlles, Dra. Míriam Barrecheguren, Dra. Cristina Esquinas Hospital Universitari Vall d’Hebrón, Barcelona, Spain; Dr. Jose Luis Izquierdo, Hospital Universitario de Guadalajara, Guadalajara, Spain; Dr. Vicente Plaza, Dr Alfons Torrego, Hospital de la Santa Creu y Sant Pau, Barcelona; Institut d’Investigació Biomédica Sant Pau (IIB Sant Pau); Universitat Autònoma de Barcelona, Department of Medicine, Barcelona, Spain; Dr. Joan B Soriano, Instituto de Investigación Hospital de la Princesa, Universidad Autónoma de Madrid, Madrid, Spain.