By analysis of a case of discrepancy between serum alpha-1-antitrypsin (AAT) level and genotype for the most common defective alleles associated with AAT deficiency (PI*S and PI*Z), a patient carrying the allele PI*Q0ourém has been identified for the first time outside of Portugal. This null allele has been implicated in cases of severe pulmonary emphysema. After developing a clinical assay for detection of c.1130insT mutation, based on fluorescent probes (HybProbe®), another 4 carriers of PI*Q0ourém allele were identified among 43 patients with abnormally low serum AAT levels based on their genotypes for PI*S and PI*Z alleles. Since 4 out of 5 cases are from the same locality (La Palma Island, Spain), it is advisable to conduct genetic analyses of affected families and, possibly, a focused population screening.
Mediante el análisis de un caso de discordancia entre el nivel de alfa-1-antitripsina (AAT) en suero y el genotipo para los alelos deficientes más frecuentemente asociados al déficit de AAT (PI*S y PI*Z), se ha identificado por primera vez fuera de Portugal un paciente que presenta el alelo nulo PI*Q0ourém, el cual ha sido asociado a casos graves de enfisema pulmonar. La puesta a punto de un ensayo clínico para la detección de la mutación c.1130insT, basado en sondas fluorescentes de tipo HybProbe®, ha permitido detectar otros 4 sujetos portadores del alelo PI*Q0ourém entre un conjunto de 43 pacientes que mostraban niveles séricos de AAT anormalmente bajos atendiendo a su genotipo para los alelos PI*S y PI*Z. Puesto que 4 de los 5 casos se concentran en una misma localidad de la isla de La Palma (España), es aconsejable realizar estudios genéticos familiares y quizás un cribado poblacional localizado.
Alpha-1 antitrypsin deficiency (AATD) is one of the most common hereditary disorders in Caucasian populations. Serum alpha-1 antitrypsin (AAT) levels of less than 80mg/dl, measured by nephelometry, increase the risk of pulmonary emphysema in adults.1 The most common deficiency variants in the SERPINA1 gene that encodes AAT are the PI*S and PI*Z alleles, found in 10.4% and 1.7% of the Spanish population, respectively.1 Ninety-five percent of the clinical cases related to AATD are associated with the PI*ZZ genotype, while the remaining 5% are associated with the PI*SZ or PI*MZ genotypes, or combinations of PI*S or PI*Z with other extremely rare deficient or null alleles.2 These rare alleles account for 1.6% of the deleterious variants recorded in the Spanish Registry of Patients with AAT Deficiency.3 Null mutations compromise the stability of the messenger RNA or the protein, resulting in extremely low or undetectable serum AAT levels.2 Therefore, genotypes composed of homozygous null alleles or accompanied by other SERPINA1 deficiency alleles have a particularly high risk of very early onset pulmonary emphysema.4 We present the case of a patient diagnosed with AATD, who was found to be a carrier of the null allele PI*Q0ourém. This is the first time that this allele has been identified outside Portugal.
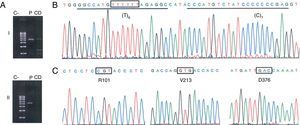
Clinical FindingsCase 1A 38-year-old man, native of La Palma Island (Spain), with no toxic habits or occupational exposure to respiratory irritants, was seen in the Respiratory Medicine clinic for persistent cough following a respiratory infection, accompanied by wheezing and dyspnea. Initial tests, including lung function tests, chest X-ray and laboratory tests, were normal. He was prescribed inhaled corticosteroids, but due to the persistence of symptoms, it was decided to perform further studies. These included immunology tests (anti-nuclear antibodies [ANA], extractable nuclear antigens [ENA] and anti-neutrophil cytoplasmic antibodies [ANCA]), high resolution computed axial tomography, the walk test, and nephelometric measurement of serum AAT concentrations. The latter was the only test to show an abnormal result, with an AAT concentration of 38.6mg/dl (reference value: 100–180),1 so the sample was analyzed for the presence of PI*S and PI*Z alleles. The patient's genotype (PI*MS; M=non-S/non-Z) was not consistent with the notably low serum AAT level. In view of these results, molecular analysis of the SERPINA1 gene was performed, focusing on the coding region of the PI*non-S allele using allele-specific polymerase chain reaction (PCR) and sequencing. This analysis revealed the presence of the PI*Q0ourém allele (Fig. 1). To our knowledge, this is the first time that this allele has been described outside Portugal. This null allele, described by Seixas et al.,5 is characterized by the insertion of a thymine nucleotide in the coding region of exon 5 within a small microsatellite (T)5, on an M3 normal background. The resulting frameshift creates a premature stop codon that shortens the carboxy-terminal end of the 19-amino acid protein, including a proline residue that is essential for AAT secretion. The mutation, while not affecting the size or stability of the messenger RNA, causes the altered protein to be retained in the endoplasmic reticulum and degraded.5
Identification of the null allele PI*Q0ourém. (A) Selective amplification of the PI*non-S allele in overlapping amplicons that cover the entire coding region of the SERPINA1 gene (NC_000014) and the corresponding introns. Fragment i (2458 bp; upstream of the non-S site) was amplified with the primer pair TACTTGGCACAGGCTGGTTT//TACTTGGCACAGGCTGGTTT, and fragment ii (2580 bp; downstream of the non-S site) with the GGGAAACTACAGCACCTGGA//GGCAGGGACCAGCTCAAC pair. Position 3′ of the allele-specific primer that discriminates the PI*S allele is highlighted in bold; this required optimization of the annealing temperature during the PCR (70°C) (C: negative control; CD: discrimination control, consisting of gDNA from an individual with the PI*SS genotype; P: gDNA from the patient in case 1). (B, C) Sequencing electropherograms for the PI*non-S allele amplified by allele-specific PCR. The site affected by the insertion of a thymine nucleotide is indicated by a box; the binding sequences of the probes designed for detection of this mutation are underlined. C shows the codons and amino acids that define the M3 genetic background.2
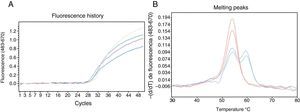
After developing an assay for detecting the null mutation involved in generating the PI*QOourém allele (Fig. 2), the method was applied to genotype the PI*S and PI*Z alleles in a group of patients from the Respiratory Medicine clinic whose serum AAT levels were below the reference range.1 The test was performed on 43 subjects, including 13 PI*MM, 28 PI*MS and 2 PI*MZ, and led to the identification of 4 new carriers of mutation c.1130insT; the presence of the M3 background was confirmed by sequencing. These cases were 4 women with normal lung and liver function. Their genotypes, serum AAT level (nephelometry), age, respiratory disease symptoms and smoking habits (respectively) were as follows: Case 2: Pi*MQ0ourém, 92.1mg/dl, 32-year-old, asymptomatic, smoker; Case 3: Pi*MQ0ourém, 70.5mg/dl, 58-year-old, asymptomatic, non-smoker; Case 4: Pi*MQ0ourém, 65.8mg/dl, 75-year-old, bronchial asthma, non-smoker; and Case 5: Pi*ZQ0ourém, 14.5mg/dl, 57-year-old, asymptomatic, non-smoker.
Detection of mutation c.1130insT in the SERPINA1 gene using HybProbe® probes. (A) Real-time monitoring of the amplification process of a 384 bp DNA fragment. (B) Results of the probe/amplicon duplex melting analysis obtained with gDNA samples from 4 patients: 2 homozygous for the wild-type variant (red) and 2 heterozygous carriers of the c.1130insT mutation (blue). The oligonucleotides used were: CGCTTCCTGGGAGGTGT (direct primer), TGGGAGGGATTTACAGTCACA (reverse primer), GGCCATGTTTTTTAGAGGC (sensor probe; 3′-fluorescein) and CCCATGTCTATCCCCCCCGAG (anchor probe; 5′-Cy5 and 3′-phosphate). The reaction mixture (20μl) contained the gDNA sample, 0.2μM of each of the 4 oligonucleotides mentioned, 200μM of each dNTP, 3mM of MgCl2, 1× reaction buffer and 0.2μl of DNA polymerase (Phire® II, Finnzymes). The reactions were incubated in the Roche LightCycler®480 system. After initial denaturation at 98°C for 30s, 50 amplification cycles with the following thermal profile were applied: denaturation, 98°C, 5s; oligonucleotide annealing, 50°C, 5s; primer extension, 72°C, 10s. The amplification was monitored by fluorescence capture (excitation at 483nm and detection at 670nm) at the end of each annealing step. After the amplification, the melting analysis was carried out, which consisted of: 95°C, 1min; 30°C, 1.5min; gradual heating up to 80°C, with 3 fluorescence captures per °C. The melting curve data were processed using the genotyping program installed in the analysis platform.
Four of the 5 carriers of the PI*Q0ourém allele detected in this study are from the same small town. Their presence on La Palma Island is probably due to the large number of Portuguese that colonized the Canary Islands. Given the severity of pulmonary emphysema symptoms associated with genotypes homozygous for the null allele PI*Q0ourém,6 a screening program to identify high risk subjects should be carried out in order to urge them to adopt healthy lifestyles and to offer genetic counseling. In this respect, an assay capable of detecting the most common rare deficient alleles in each population is very useful.3 The genotyping method described in this article is simple and inexpensive, allowing good resolution of the mutant and wild-type variants (approximately 5.5°C). As the sensor probe is fully complementary to the mutant variant, this will always produce the curve with the highest melting temperature, identified with total confidence within a PI*Q0ourém or PI*Q0mattawa allele (M1-Val213 genetic background). It should be noted that the anchor probe covers a mutation hotspot (microsatellite (C)7, Fig. 2); therefore, the assay could potentially detect other null alleles such as PI*Q0clayton (c. 1158insC in M1-Val213), PI*Q0saarbruecken (c. 1158insC in M1-Ala213) or PI*Q0bolton (c. 1158delC in M1-Val213). In this case, the mutant allele would have a lower melting peak than the wild-type variant in the genotyping assay.
FundingGrifols International S.A.
Conflict of InterestThe authors state that they have no conflicts of interest directly or indirectly related with the contents of the manuscript.
The authors would like to thank Grifols International S.A. for funding this study.
Please cite this article as: Hernández Pérez JM, Ramos Díaz R, Fumero García S, Pérez Pérez JA. Descripción de la deficiencia de alfa-1-antitripsina asociada al alelo PI*Q0ourém en la isla de La Palma (España) y de un método de genotipado para su detección. Arch Bronconeumol. 2015;51:e1–e3.