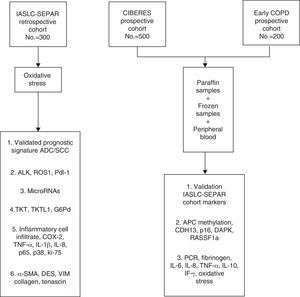
The aim of the Clinical and Molecular Staging of Stage I-IIp Lung Cancer Project is to identify molecular variables that improve the prognostic and predictive accuracy of TMN classification in stage I/IIp non-small cell lung cancer (NSCLC). Clinical data and lung tissue, tumor and blood samples will be collected from 3 patient cohorts created for this purpose. The prognostic protein signature will be validated from these samples, and micro-RNA, ALK, Ros1, Pdl-1, and TKT, TKTL1 y G6PD expression will be analyzed. Tissue inflammatory markers and stromal cell markers will also be analyzed. Methylation of p16, DAPK, RASSF1a, APC, and CDH13 genes in the tissue samples will be determined, and inflammatory markers in peripheral blood will be analyzed. Variables that improve the prognostic and predictive accuracy of TNM in NSCLC by molecular staging may be identified from this extensive analytical panel.
El proyecto «Estadificación clínica y molecular del cáncer de pulmón estadios I-IIp», tiene por objetivo la identificación de variables moleculares que mejoren la capacidad pronóstica y predictiva de la clasificación TNM en el cáncer de pulmón no célula pequeña (NSCLC) estadio I/IIp. Para ello se han creado tres cohortes de pacientes de los que se recoge información clínica y muestras biológicas pulmonares y de sangre. En las muestras se validará una firma pronóstica proteica, y se analizarán micro-RNA, ALK, Ros1, Pdl-1, y niveles de expresión de TKT, TKTL1 y G6PD. Asimismo se analizarán marcadores de inflamación en tejido y marcadores de estroma. Se determinará la metilación de los genes p16, DAPK, RASSF1a, APC y CDH13 en la muestra tisular y se analizarán marcadores inflamatorios en sangre periférica. Este amplio panel analítico puede permitir la identificación de variables que mejoren la capacidad pronóstica y predictiva del TNM en el NSCLC mediante la estadificación molecular.
Lung cancer (LC) is the leading cause of cancer-associated deaths,1,2 and has a 5-year survival rate of less than 18%.3 Only very few cases are diagnosed at the limited stage of the disease, when surgical resection is indicated.4 Early detection of cancer improves prognosis; however, this is hampered by the absence of symptoms.3 Approximately 80% of all lung cancer patients are diagnosed with non-small cell cancer (NSCLC),5 adenocarcinoma and squamous cell carcinoma being the most common subtypes. For the purpose of prognosis, lung cancer must be staged according to the extent of the primary tumor (T), lymph node involvement (N), and metastasis (M), although comorbidity must also be taken into account.4
The identification of predictive or prognostic tumor factors such as epidermal growth factor receptor mutations,6–11 together with translocation of the EML4-ALK (Anaplastic Large Cell Lymphoma) gene12–14 has contributed to the exponential growth in the number of LC tumor markers identified and developed over the past decade. The priority now is to add prognostic and predictive factors to the TNM staging system to identify patient subgroups that are candidates for targeted cancer therapies.6,7,15–17 For a significant number of markers, however, findings have so far been inconclusive. The slow progress in obtaining clinically relevant results is due as much to the complexity of the disease as to the analytical methodology used.4
The Clinical and Molecular Staging of Stage I-IIp Lung Cancer Project is part of the Strategic Project in Lung Cancer, organized by the Biomedical Research Networking Center (CIBERES) in conjunction with the Spanish Society of Pulmonology and Thoracic Surgery (SEPAR) and the Thematic Network of Collaborative Research in Cancer (RTICC). The aim of the project is to collect clinical data, lung tissue, tumor tissue and blood samples from a cohort of patients with limited-stage (I-IIp) NSCLC receiving surgical treatment. The research groups taking part in the study will analyze potentially prognostic and/or predictive genetic, epigenetic and proteomic markers. This information will then be combined with the clinical characteristics of each patient to give a multidimensional classification of stage I/IIp NSCLC, in which factors contributing to the anatomical extension of the tumor are just one component in a validated panel of prognostic and predictive factors.
The data and samples will be collected from 3 cohorts of stage I/IIp cancer patients that have undergone surgery. The first cohort, the retrospective International Association for the Study of Lung Cancer (IASLC) series, was formed from the registry of clinical data and paraffin-embedded tissue samples collected by SEPAR's Lung Cancer Cooperative Group (GCCP-II).18 The second prospective cohort, the CIBERES series, includes new cases of stage I/IIp NSCLC. Clinical data from this group are recorded in a registry and tumor tissue, healthy lung tissue, and peripheral blood samples are stored in a biobank for subsequent molecular analysis. The third cohort formed for this project is the Early COPD series, which comprises cases of NSCLC identified from a screening program in smokers aged over 45 years with chronic obstructive lung disease (COPD).
The main aim of the project is to identify clinical and molecular variables that can improve the prognostic and predictive accuracy of the TNM I/IIp cancer staging system. In this article, we will describe the cohort selection criteria, the baseline and follow-up clinical data obtained, the methodology used for processing this information and for storing samples in the CIBERES Pulmonary Biobank Consortium (PBP), and analyze the procedure used (Fig. 1).
MethodDesign, Population and Clinical VariablesThis is a consecutive cohort study lasting 2 years, in which patients with stage I/IIp NSCLC will be recruited at the time of their surgery. All patients will agree to take part in the study and for their samples to be stored in a biobank. We hope to recruit a total of 1000 patients from all 3 study cohorts.
Clinical data from each patient will be collected at the time of inclusion, and the 5-year follow-up period started. Variables from the IASLC TNM cancer staging project18,19 will be recorded, namely sociodemographic data, smoking history, comorbidities, blood work, lung function, tumor site, tumor type and differentiation, staging studies (computed and positron emission tomography, bone scan, brain CT and/or MRI, endoscopy and/or mediastinoscopy), surgical resection and node dissection, adjuvant treatment, and a detailed description of the TNM descriptors.20,21 For follow-up, information on recurrence and mortality will be collected: (a) disease-free interval; (b) recurrence; (c) survival; and (d) cause of death (see Supplementary information).
Sampling and Storage in the Pulmonary Biobank ConsortiumPathology samples of tumor tissue, healthy lung tissue and blood are processed immediately following extraction according to Pulmonary Biobank Consortium (PBP) protocols22 (Appendix B, Table 1, Supplementary information). The PBP is in charge of organizing and overseeing the sampling of lung tissue and other related samples and linking these with the clinical information, and ensuring they are readily accessible to study investigators.23 The PBP is a branch of the Spanish National Biobanks Consortium, made up of Spanish hospitals that voluntarily contribute cases and samples. The sample collection period for all 3 cohorts will be 2 years, and samples will have been analyzed within 1 year after the end of the recruitment period. Non-paraffinized tissue samples and peripheral blood samples are stored at −80°C, and analyzed within 1 year.24
Preliminary Processing of Paraffin-Embedded SamplesParaffin-embedded tissue samples are collected from all 3 study cohorts. In cohorts with hematoxylin and eosin (H&E) stained samples, the most homogeneous tumor areas in each block are chosen to obtain three, 0.1cm diameter cylinders which are used to create independent tissue microarrays for immunohistochemical (IHC) analysis of each histological NSCLC subtype. Whenever possible, a fourth cylinder of the same size is created from healthy tissue adjacent to the tumor, and a fifth from transitional tissue. These are used for separate microarrays.
After macrodissection of the paraffin-embedded samples to obtain the greatest possible proportion of tumor cells, further sectioning is performed to extract nucleic acid. While the microarrays are being prepared, stained samples are evaluated by a panel of pathologists by means of a semiquantitative assessment of tissue differentiation and vascular, nodal and adjacent tissue involvement.
Biological VariablesParaffin-Embedded Samples(a) Validation of a prognostic protein signature
Prognostic signatures for NSCLC were developed from microarray-based mRNA expression profiling.25–31 These profiles are difficult to translate and reproduce in the clinical setting. However, this problem can be overcome by translating these signatures to their corresponding prognostic protein signature, using IHC to quantify their expression levels in paraffin-embedded tissue samples. The project will develop a protein signature capable of classifying early-stage NSCLC patients into subgroups according to their risk for recurrence.
Based on the findings of earlier studies, we have selected a group of 15–20 proteins that can be used for classifying early-stage NSCLC patients into 2 groups, according to their clinical course. For this purpose, we used an algorithm generated by the biomarker laboratory of the Center for Applied Medical Research of the University of Navarre. The proteins selected include a panel of 5 RNA binding proteins that have been shown to be prognostic markers in cohorts of patients with early-stage NSCLC.32 On this basis, we were able to generate a prognostic protein signature that will be validated in our study cohorts.
(b) ALK, Ros 1 and Pdl-1 determinations
ALK immunohistochemistry assay using two different, validated antibodies, D5F3 and 5a4, will be performed to study the correlation between these genes. All positive results will be confirmed using fluorescence in situ hybridization. IHC study of Ros-1 will be performed to ascertain the percentage of molecular changes. The same technique will be used to determine Pdl-1 and its relationship with survival. Previous studies in Pdl-1 in LC have been based on a limited number of cases; however, studies of Pdl-1 expression in mesothelioma reported a relationship between Pdl-1 expression and patient survival.
(c) Micro-RNA
Micro-RNA (miRNA) are non-coding molecules of 19–25 nucleotides in length. Their function is to regulate the post-transcriptional expression of multiple genes. They are involved in many different cell cycle processes,33 and have been shown to play a leading role in cancer progression.34,35 Tumor-derived miRNAs can enter the bloodstream,36 and their expression profile in serum and plasma is disease-specific.37 Because of this, and because they are stable, easily detectable molecules, miRNAs are now the subject of intense study.38,39 In this project, we will select miRNA to be studied on the basis of the following criteria:
- (1)
miRNAs that have been identified in at least 3 published studies as potential biomarkers of NSCLC prognosis.
- (2)
EGFR-, ALK- and KRAS-regulating miRNAs, validated in functional studies, retrieved from a search of a database of miRNA target prediction resources.
- (3)
miRNAs that regulate other genes of interest in the context of NSCLS that have been studied by researchers taking part in the study (see Supplementary information).
(d) Pentose phosphate pathways
The pentose phosphate pathway plays an important role in the development of cancer because it generates the nitrogenous base needed to synthesize DNA. The non-oxidative phase of the pathway also synthesizes nicotinamide adenine dinucleotide phosphate, which is essential in the synthesis of the fatty acids needed to form cell membranes.
Because of its importance in biomolecular synthesis and the proliferation of the tumor itself, this pathway is regulated by some of the most important proto-oncogenes and tumor-suppressor genes. Research has shown that tumors with a mutated KRAS gene present increased levels of glucose-6-phosphate dehydrogenase (G6PD),40 and cell proliferation via this molecule is directly related with proteins such as TAp73.41 G6PD inhibition can slow down tumor growth,42 while more aggressive tumor growth has been correlated with over-expression of TKTL1.43 In the specific case of LC, recent studies suggest that both TKT and G6PD can be over-expressed,44 which justifies the interest of researchers in pentose phosphate enzymes as potential biomarkers of LC (see Supplementary information).
(e) Inflammatory markers
Chronic inflammation plays a key role in the pathogenesis of NSCLC, particularly in patients with COPD.45,46 Experimental studies have shown increased expression of certain cytokines such as interleukin (IL)-6, IL-8, and IL-10 in patients with COPD and a history of smoking. These proteins promote the lymphocytic inflammatory response by induction of the cyclooxygenase-2 enzyme. These cytokines can inhibit apoptosis, interfere with cell repair mechanisms, and encourage angiogenesis. Other inflammatory mechanisms implicated in LC are mediated by the angiogenesis-regulating vascular endothelial growth factor, the epidermal growth factor, the tumor necrosis factor (TNF)-alpha, the nuclear factor (NF)-kB, and the tumor growth factor-beta (TGF-beta). Chronic inflammation can be important in amplifying mutagenesis, facilitating tumor growth, and increasing the frequency of metastasis.
In this project, IHC assay will be used to quantify the following markers:
- (1)
Inflammatory cell infiltrate in H&E-stained histological samples.
- (2)
Specific cell infiltrate, using specific antibodies to identify leukocytes (anti-CD45) and macrophages (anti-CD68).
- (3)
Levels of inflammatory molecules involved in tumor development processes:
- (3.1)
Cyclooxygenase-2 (COX-2), involved in angiogenesis, tumor invasion, and immune system suppression.
- (3.2)
TNF-alpha, a molecule that induces COX-2 expression through mitogen-activated protein kinases (MAPKs) signaling pathways.
- (3.3)
IL-1beta and IL-8, which induce COX-2 expression and are involved in neutrophil recruitment and tumor progression.
- (4)
NSCLC-related signaling pathways, such as p65 (NF-kB) and p38 (MAPK).
- (5)
Determination of the extent of cell proliferation in tumors by quantifying nuclear protein ki-67 expression as a specific marker of tissue growth (see Supplementary information).
(f) Stromal cell activity markers
An important signaling source for cancer cells is stromal change, characterized by an over-abundance of extracellular matrix components and activated fibroblasts, and by abnormal composition of the extracellular matrix. Stromal changes are also observed in fibrogenetic processes. It is important to identify the extent to which signals from activated stroma contribute to LC progression, as this can guide therapeutic strategies. Changes in tumor stroma have been studied in the context of breast cancer, but little is known about their association with LC. Since myofibroblasts are the most abundant cells in activated stroma, we will use IHC to study alpha smooth muscle actin expression (α-SMA). To distinguish myofibroblasts from other cells, we will examine desmin and vimentin expression,47 and the presence of extracellular matrix cicatrizing components will be determined by analyzing collagen and tenascin expression48 (see Supplementary information).
Frozen SamplesDNA methylation helps regulate gene expression, and studies have shown that aberrant DNA methylation is frequently found in malignancies. Methylation is found in CpG islands, which are concentrated in promoter regions and in the first exon of protein-coding genes. Up to 60% of these genes contain CpG islands and their promoter sequences, and their methylated status is inversely correlated with transcription.49
The following genes: p 16 (cyclin-dependent kinase inhibitor 2) (CDKN2A/p16), H-cadherin (CDH13), adenomatous polyposis coli (APC), Ras association domain family 1 gene (RASSF1A), and death-associated protein kinase 1 (DAPK) are important in the development of NSCLC, and are frequently methylated in tumor tissue.50,51 Methylation of p16, DAPK, CDH13, APC and RASSF1A has been associated with an increased risk from LC,52 and early recurrence.51 It could be important to determine the methylation status of these genes for both diagnosis and prognosis (see Supplementary information).
Peripheral Blood SamplesThe characteristics of the inflammatory response in peripheral blood cells, which is partly mediated by the generation of oxidative stress,53 can affect prognosis and therapeutic response in LC. Cancer-related inflammation is also a new therapeutic target,54 and various LC-related inflammatory markers in blood have been described. In this project, we will measure circulating levels of acute phase reactants, inflammatory cytokines, and markers of oxidative stress.
- (1)
Acute phase reactants:
- (1.1)
C-reactive protein is a non-specific reactant associated with LC through various mechanisms, including tumor-induced inflammatory response, immune response to tumor antigens, and cytokine production by the tumor cells themselves. High levels of C-reactive protein are predictors of mortality.55
- (1.2)
Fibrinogen has been associated with increased risk for LC.56
- (2)
Inflammatory cytokines:
- (2.1)
IL-6, IL-8 and TNF-α can be elevated in LC due to mechanisms similar to those described for C-reactive protein, and increased circulating levels of these cytokines have been associated with LC.57
- (2.2)
Over-expression of the inflammatory cytokine IL-10 has been associated with CP progression.58
- (2.3)
Interferon gamma deficiency has been associated with LC in experimental models,59 and in human series.60
- (3)
Oxidative stress:
Circulating oxidative stress has been associated with risk for LC,61 advanced disease,62 and therapeutic response63 (see Supplementary information).
The clinical and follow-up information obtained from all 3 NSCLC stage I/IIp cohorts will first be used to perform a general and comparative description of cohorts, following which all the available blood and tissue samples will be analyzed for biomarkers. Underlying patterns will be examined using computational biology techniques.
We will study the association between study outcomes (recurrence and mortality) and variables: (a) patient clinical characterization variables (sociodemographic, smoking history, comorbidities, blood work, lung function), (b) tumor-related variables (site, type and differentiation), (c) staging variables, (d) surgical treatment (characteristics and node dissection), (e) adjuvant therapy, (f) TNM, and (g) biological variables.
Clinically relevant variables that have a documented association in earlier univariate analyses will be selected for use in multivariate models. Multivariate logistic regression models will be used for each outcome, and Cox regression will be used to study changes in survival curves on the basis of the explanatory variables retained in each model.
Finally, the models created from the findings from the retrospective IASLC cohort will be validated in the CIBERES cohort and the Early COPD cohort.
FundingThis study has been sponsored by the CIBER of Respiratory Diseases (CIBERES), an initiative launched by the Instituto de Salud Carlos III, the Spanish Society of Pulmonology and Thoracic Surgery (SEPAR), the Thematic Network of Collaborative Research in Cancer (RTICC), the Spanish Health Research Fund (FIS PI02040, PI01310 and PI02534) and the Spanish Cancer Association (AECC).
Conflict of InterestThe authors declare they have no conflict of interest.
CIBERES–RTICC–SEPAR Lung Cancer Group: Jordi Alcaraz,2,6 Carlos Álvarez,2,7 Pilar Ausin,2,8 Joan Albert Barberà,2,19 Esther Barreiro,2,8 Ana Isabel Blanco,9 Anna Brunet,1°,11 Marta Cascante,2,6 Ady Castro,2,7 José Ignacio Echeveste,12 Ana Belén Enguita,2,13 Leonardo de Esteban Júlvez,14 Esther Fernández,15 M. Jesús Fernández-Aceñero,16 M. Ángeles González Castro,9 Guadalupe González Pont,17 Pedro López de Castro,2,15 María Dolores Lozano,12 Ramón Marrades,18 Carlos Martínez-Barenys,15 Elena Martínez-Terroba,3,4 José Luis Mate,20 Rosario Melchor,16 Laura Millares,2,10 Eduard Monsó1,2 (CIBERES coordinator), Luis M. Montuenga3,4 (RTICC coordinator), Mariano Monzó,19 Alfons Navarro,19 María José Pajares,3,4 Carles Pericay,11 Lara Pijuan,21 Ramón Rami-Porta,2,17 Josep Ramírez,2,18 Antoni Rosell,2,22 Julio Sánchez de Cos2,5 (SEPAR coordinator), Rosa María Sánchez Gil,9 Jaume Sauleda,2,23,24 Sergio Scrimini,2,23,24 Luis Seijo,2,16 Mireia Serra,17 Laia Setó2 and Cristina Villena.2
- (1)
Department of Respiratory Medicine, Hospital Universitari Parc Taulí, Sabadell.
- (2)
CIBER of Respiratory Diseases–CIBERES, Instituto de Salud Carlos III, Madrid.
- (3)
Solid Tumor Pathogenesis Program, Biomarker Laboratory, Center for Applied Medical Research (CIMA), University of Navarre RTICC Group RD12/0036/0040, Pamplona, Spain.
- (4)
Department of Histology and Anatomical Pathology, Faculty of Medicien and Facultiy of Science, University of Navarre, Pamplona, Spain.
- (5)
Department of Respiratory Medicine, Hospital San Pedro de Alcántara, Cáceres, Spain.
- (6)
University of Barcelona, Barcelona, Spain.
- (7)
Department of Respiratory Medicine–Research Institute, Hospital Universitario 12 de Octubre, Madrid, Spain.
- (8)
Department of Respiratory Medicine–Lung Cancer Research Group, Hospital del Mar–IMIM, Department of Experimental and Health Science, Universidad Pompeu Fabra, Parque de Investigación Biomédica de Barcelona (PRBB), Barcelona, Spain.
- (9)
Hospital Virgen del Rocío, Seville, Spain
- (10)
Fundació Parc Taulí, Sabadell, Spain.
- (11)
Department of Medical Oncology, Hospital Universitari Parc Taulí, Sabadell, Spain.
- (12)
Department of Anatomical Pathology, Clínica Universidad de Navarra, RTICC Group RD12/0036/0062, Pamplona, Spain.
- (13)
Department of Anatomical Pathology - Research Institute, Hospital Universitario 12 de Octubre, Madrid, Spain.
- (14)
Hospital Joan XXIII, Tarragona, Spain.
- (15)
Department of Thoracic Surgery, Hospital Universitari Germans Trias i Pujol, Badalona, Spain.
- (16)
Fundación Jiménez Díaz, Madrid, Spain 17. Hospital Mutua de Terrassa, Terrassa, Spain.
- (17)
Hospital Clínic, Barcelona, Spain.
- (18)
Hospital Clínic–Faculty of Medicine, University of Barcelona, Barcelona, Spain.
- (19)
Anatomical Pathology, Hospital Universitari Germans Trias i Pujol, Badalona, Spain.
- (20)
Pathology Department, Hospital del Mar, Barcelona, Spain.
- (21)
Department of Respiratory Medicine, Hospital Universitari de Bellvitge, L’Hospitalet de LLobregat, Spain.
- (22)
Department of Respiratory Medicine, Hospital Universitari Son Espases, Palma, Spain.
- (23)
Health Research Institute of Palma, IdISPa.
A list of members of the Lung Cancer CIBERES-RTICC-SEPAR-Plataforma Biobanco Pulmonar can be found in Appendix A.
Please cite this article as: Monsó E, Montuenga LM, Sánchez de Cos J, Villena C, por el Grupo Colaborativo en Cáncer de Pulmón CIBERES-RTICC-SEPAR-Plataforma Biobanco Pulmonar. Análisis de marcadores biológicos en el Proyecto Estratégico de Cáncer de Pulmón CIBERES-RTIC Cáncer-SEPAR. Arch Bronconeumol. 2015;51:462–467.