The respiratory system is constantly being exposed to airborne pathogens. Thus, it is not surprising that respiratory tract infections (RTIs) are the most prevalent acute diseases in primary care and among one of the top causes of death globally each year.1,2 Bacteria are one of the main pathogens responsible for primary and secondary RTIs. Recently, secondary bacterial RTIs have been brought to the forefront due to the COVID-19 pandemic. Numerous patients infected with SARS-CoV-2 present RTIs caused by “superbugs”, which substantially worsen clinical outcomes.3 RTIs are commonly treated using antibiotics.2,4 However, antibiotics are increasingly losing their efficacy against bacteria due to the emergence of antimicrobial resistance (AMR). Priority pathogens that infect the respiratory system may develop resistance, leading to severe and recalcitrant RTIs. Antibiotics traditionally used to treat such infections are broad-spectrum, causing off-target effects on the beneficial lung microbiota.5 The future of pulmonary infection treatment may involve developing antimicrobials that target pathogens while leaving unaffected our beneficial microbes.
Antimicrobial peptides (AMPs) are a promising class of antimicrobial candidates that are naturally produced and serve as the primary defence mechanism of many organisms against pathogenic bacteria, viruses, parasites, and even cancer cells (Fig. 1).6 AMPs are commonly short, amphiphilic, and cationic molecules with broad-spectrum activity.7 These peptides can attack bacteria by compromising their membrane integrity through non-specific mechanisms of action (MoAs),8 can modulate the host's immune response to clear infections, or even balance potentially harmful inflammation.7,9 The ability of these agents to target highly drug-resistant bacteria while not selecting for bacterial resistance confers advantages over classical antimicrobials.
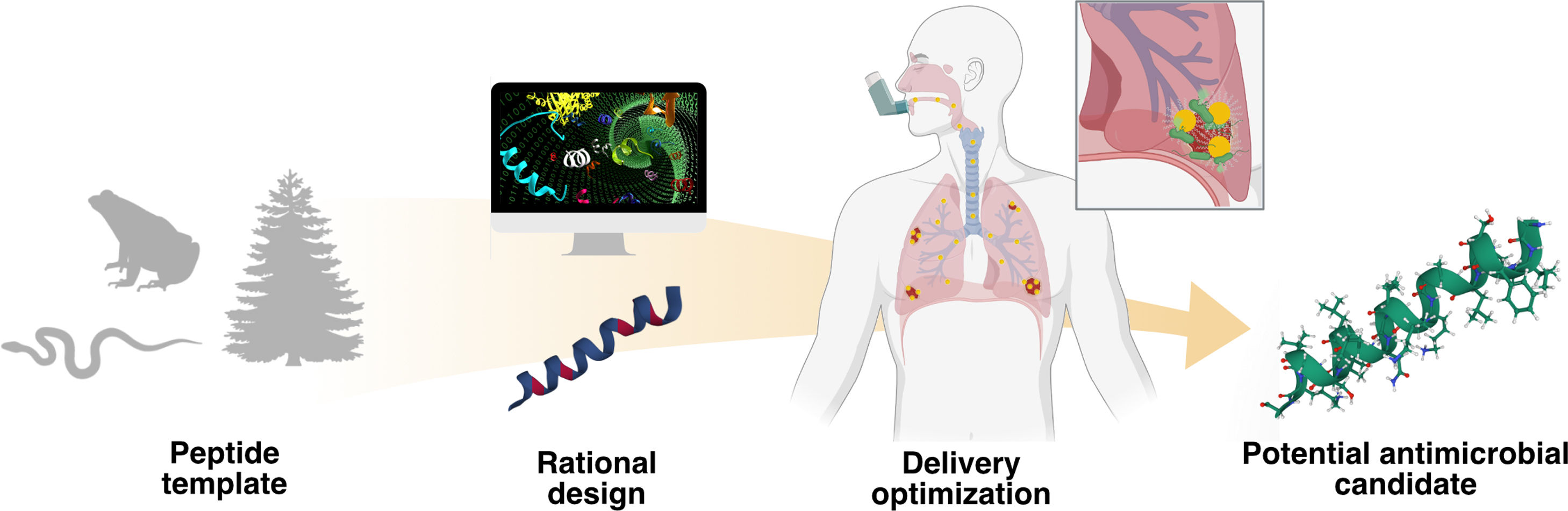
Schematic representation of the AMP optimization process against “superbugs” responsible for RTIs. Nature provides multiple peptide templates that can be rationally designed through different methodologies, such as site-directed mutagenesis or computational-based approaches, yielding potential lead molecules. Due to the difficulty of the administration of AMPs on the lungs, different delivery optimization approaches should be done in order to obtain a potential antimicrobial candidate.
Several studies have examined the efficacy, versatility, and proper delivery of natural AMPs against “superbugs” responsible for RTIs. For example, Esc(1–21) (GIFSKLAGKKIKNLLISGLKG-NH2), the N-terminal fragment of the natural peptide Esculetin-1a, has demonstrated excellent in vitro antimicrobial activity (MIC<8μmolL−1) against multi-drug resistant and cystic fibrosis clinical isolates of Pseudomonas aeruginosa, and prolonged survival rates in sepsis and lung infection in animal models.10 This peptide also repressed the re-growth of bacteria on pre-treated biofilms (MBEC=8μmolL−1) and eliminated biofilm cells within 2h.10
The versatility of the peptide backbone makes it amenable for reprogramming in order to enhance a given biological activity (Fig. 1). For instance, l-amino acids can be substituted by d-amino acids to reduce degradation by proteases and to improve stability and antimicrobial activity.11 Di Grazia et al. performed a mutagenesis study using peptide Esc (1–21), yielding variants with significantly reduced cytotoxicity towards mammalian cells, improved anti-biofilm activity, peptide biostability, and increased promotion of lung epithelial cells migration.12 As outlined in the review by Torres et al., many peptide design approaches rely on structure-activity relationships (SAR), which include site-directed mutagenesis, computational design approaches, synthetic libraries, template-assisted methodologies, and mechanism-based strategies.7,13 Site-directed mutagenesis based on physicochemical-guided and structure–function-guided peptide design are effective strategies for peptide optimization.7 Following these methodologies, our research group successfully redesigned Polybia-CP, a wasp venom peptide, into a potent synthetic derivative active at nanomolar concentrations in vitro that displayed anti-infective activity against P. aeruginosain vivo.14 Although substantial progress has been made through experimental design and validation, the functional determinants of peptides have not yet been fully elucidated. Computer-guided peptide design allow us to consider additional descriptors, vectors that describe the peptide's features, and enables exploration of novel regions of combinatorial sequence space. Computational tools are greatly expanding our ability to understand and optimize AMPs. For example, these tools have been used to predict peptide's activity,7 generate new active candidates,13 and explain biological processes, such as the MoAs of AMPs.15 The algorithm Joker, based on the linguistic model approach, inserts antimicrobial patterns in sequence templates, enhancing the activity of active peptides and creating AMPs from inactive ones.16 This algorithm yielded the synthetic peptide PaDBS1R6, which reduced the bacterial viability in vitro and in vivo against Gram-negative bacteria such as P. aeruginosa.17
Peptides may also be reprogrammed to endow them with narrow-spectrum activity towards select pathogens by rational design, computational design, and high-throughput screening.18 Among rational design approaches, species-selective peptide nucleic acids (PNA) have been developed to target specific pathogens based on their nucleic acid sequence.19 Another strategy includes linking a bacterial recognition domain with an AMP sequence, such as the enterococcal pheromone cCF10 of Enterococcus faecalis that was fused to the rationally modified AMP C6 resulting in a selective peptide against E. faecalis.20 Simple mutagenesis studies of natural peptides can also yield excellent candidates with narrow-spectrum activity. A single amino acid mutation in the natural peptide RI16 led to narrow selectivity towards P. aeruginosa and highlighted the importance of the tryptophan amino acid in certain positions of the sequence.21
One of the primary hurdles for treating RTIs with peptides is their effective delivery into the lungs, particularly in the case of cystic fibrosis (CF) patients. Several delivery approaches, such as nanoformulations that can be inhaled, have been developed to deliver peptides through the mucosal and cellular barriers while preventing degradation (Fig. 1). Recently, Casciaro et al. described the development of nanoparticles for the delivery of AMPs in the pulmonary region.22 The authors formulated stable lyophilized poly(lactide-co-glycolide) (PLGA) nanoparticles (NPs) encapsulating the peptide Esc(1–21) and Esc(1–21)1c that were able to penetrate through mucus and bacterial biofilm artificial models without interacting with the major components of the lung mucus or the extracellular matrix of P. aeruginosa biofilm communities.22 These NPs demonstrated prolonged in vitro activity against P. aeruginosa and improved the in vivo activity compared to groups treated with free peptides in an acute P. aeruginosa lung infection model in mice.22
In conclusion, natural AMPs constitute a source of structurally and mechanistically diverse biologically active molecules that serve as promising alternatives to conventional antibiotics. Experimental and computational methods have been developed that allow us to redesign these molecules to tune their structure and function, leading to synthetic peptides with excellent therapeutic activity in numerous animal models of preclinical relevance. In this editorial, we provide a brief summary of this field and its promise for the effective treatment of bacterial RTIs.
Cesar de la Fuente-Nunez holds a Presidential Professorship at the University of Pennsylvania, is a recipient of the Langer Prize by the AIChE Foundation and acknowledges funding from the Institute for Diabetes, Obesity, and Metabolism, the Penn Mental Health AIDS Research Center of the University of Pennsylvania, the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM138201, and the Defense Threat Reduction Agency (DTRA; HDTRA11810041 and HDTRA1-21-1-0014). We thank Xunta de Galicia (Spain) for financing Lucía Ageitos with a fellowship (ED481A-2019/081) co-financed by ESF (European Social Fund).