Asthma is a heterogeneous syndrome with a variety of clinical phenotypes and endotypes, including type 2 (T2) inflammatory endotypes, characterised by the predominance of interleukins (IL)-4, IL-5 and IL-13, and other types in which non-eosinophilic airway inflammation or mixed inflammation with the presence of type 1 (Th1) and type 17 (Th17) cytokines are found.1 Eosinophilic asthma represents the most prevalent phenotype, accounting for approximately 84% of all asthma cases and 50% of patients with severe asthma.2,3 It is characterised by persistent airway inflammation, elevated blood and sputum eosinophil counts, recurrent exacerbations and, in some patients, reduced lung function. Some severe eosinophilic asthma (SEA) patients respond inadequately to conventional treatments, such as oral corticosteroids (OCS), and may benefit from new antibody-based therapies.2 Exosomes, ranging from 30nm to 150nm in diameter, are small extracellular vesicles (EVs) released into the extracellular microenvironment by most cell types, including eosinophils, key cells in asthma. These nanovesicles facilitate intercellular communication, either through direct cell-to-cell contact or by transporting various molecules such as nucleic acids, lipids and proteins. They therefore play a crucial role in several physiological and pathological processes, including those associated with asthma.4 However, to date, no studies have been conducted on the protein content of exosomes in relation to the different asthma phenotypes, in particular eosinophilic asthma (EA) and non-eosinophilic asthma (NEA).
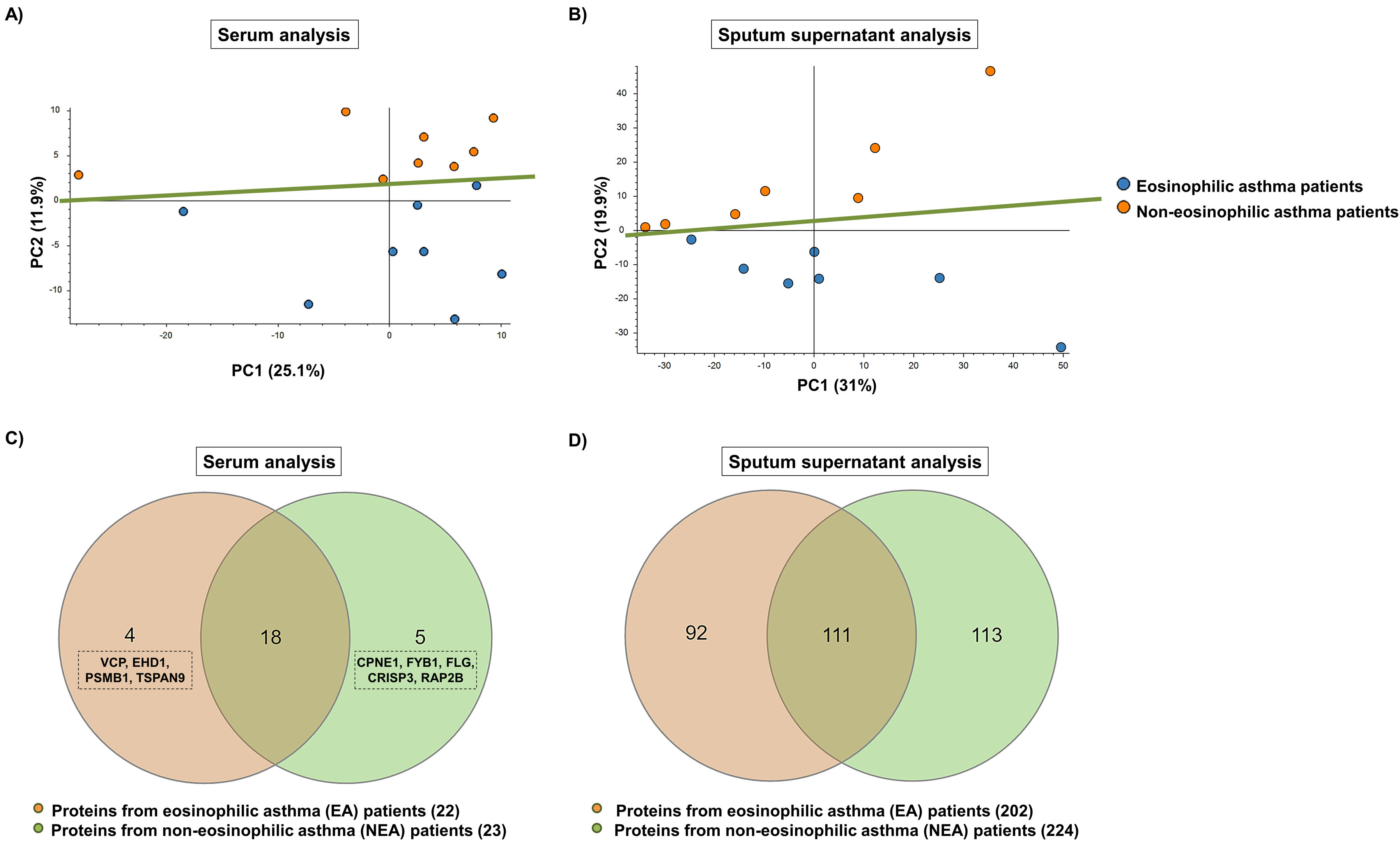
In this study, we sought to compare the protein content of serum and sputum supernatant exosomes from 15 EA and 15 NEA patients, respectively. Exosomes were first purified by ultracentrifugation from both types of samples and from the two phenotypes of asthma patients. The protein concentration of the isolated exosomes was then quantified and exosomal markers were identified by Western blotting to verify the correct purification of the exosomes. Finally, the proteomic profile was characterised by reversed-phase liquid nano-chromatography coupled to tandem mass spectrometry (RP-LC–MS/MS), followed by data analysis of relative quantification without protein labelling and in silico functional analysis of proteins with different abundance. Detailed materials and methods are provided in the Supplementary Material. Demographic and clinical characteristics of EA and NEA patients (serum and sputum supernatant analysis) are shown in Supplementary Material Table 1. In the group of patients from whom supernatant sputum samples were obtained, individuals with EA exhibited significantly higher FeNO levels than those with NEA (p<0.05). Proteome differences were evident in both analyses, thus principal component analysis (PCA), performed on the protein abundance values clearly separated the two patient groups (Fig. 1). The two principal components (PC1 and PC2) together explained 37.0% (Fig. 1A) and 50.9% (Fig. 1B) of the total variance obtained. In both cases, PC2 variances of 11.9% (Fig. 1A) and 19.9% (Fig. 1B) resulted in NEA patient samples being grouped at the top and EA patient samples at the bottom. Regarding protein content, a total of 403 proteins in serum exosomes and 1934 proteins in sputum supernatant exosomes were identified and quantified. Of these, 27 and 316 proteins, respectively, showed differences in abundance between EA and NEA patients (p<0.02 and p≤0.01) (Fig. 1). In serum exosomes, 4 proteins (VCP, EHD1, PSMB1 and TSPAN9) were unique to EA patients, 5 (CPNE1, FYB1, FLG, CRISP3 and RAP2B) were exclusive to NEA patients and 18 were common to both groups (of which 17 were more abundant in EA patients), with FLG and CD44 standing out among the 27 proteins (Fig. 1C). In sputum supernatant exosomes, 92 proteins were unique to EA patients, 113 were exclusive to NEA patients and 111 were shared by the two groups (with 77 being more abundant in EA patients), highlighting PRG2, EPX, RNASE2 and CLC among the 316 proteins (Fig. 1D).
Principal component analysis (PCA) of serum (A) and sputum supernatant (B) exosome proteins. Venn diagrams of purified exosome proteins from serum (C) and sputum supernatant (D). Proteins unique to the eosinophilic asthma (EA) and non-eosinophilic asthma (NEA) phenotypes and those common to both phenotypes are included. VCP, transitional endoplasmic reticulum ATPase; EHD1, EH domain-containing protein 1; PSMB1, proteasome subunit beta type 1; TSPAN9, tetraspanin-9; CPNE1, copine-1; FYB1, FYN-binding protein 1; FLG, filaggrin; CRISP3, cysteine-rich secretory protein 3; RAP2B, Ras-related protein Rap-2b.
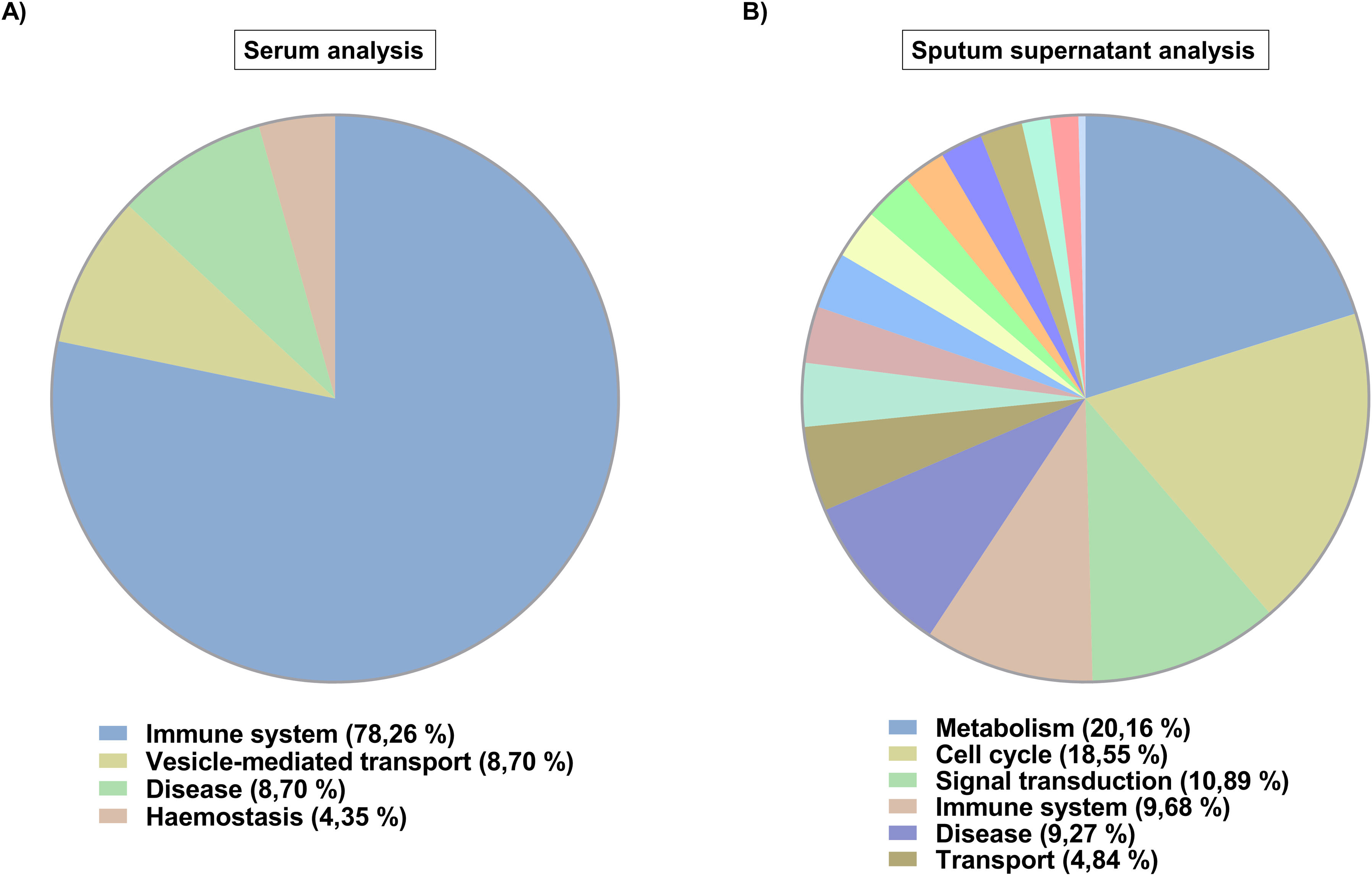
In silico analysis (Fig. 2) revealed that serum exosome proteins were involved in 23 biological processes (Fig. 2A), all of them up-regulated based on EA condition. These were distributed in four categories, most of them being related to the immune system (78.26% of the processes). On the other hand, sputum supernatant exosome proteins were implicated in 248 biological processes (Fig. 2B), 54 down-regulated and 194 up-regulated according to EA condition. The three over-regulated processes in EA with the highest number of proteins were processes related to the immune system (innate immune system and neutrophil degranulation) and genetic information processing (proteasome). The 248 biological processes were classified in 17 categories, with metabolism being the most frequent category (20.16% of the processes). Moreover, 22 biological processes involving exclusively serum exosome proteins and 247 in which only sputum supernatant exosome proteins were detected, with 1 biological process shared by exosome proteins from the two sample types: FcɛRI receptor signalling.
Advances in omics technologies have intensified studies on the protein content of EVs, with the objective of discovering effective molecular markers for disease diagnosis.5 Our study demonstrated a significant difference in the proteins identified and quantified in exosomes purified from serum (403 proteins) and sputum supernatant (1934 proteins). Among the proteins discovered by RP-LC–MS/MS, 9 out of 27 in serum exosomes and 209 out of 316 in sputum supernatant exosomes were previously reported in the complete eosinophil proteome,6 suggesting an origin of exosomes from this cell type. In serum exosomes, FLG protein was found exclusively in NEA patients, whereas CD44 was more abundant in EA patients, both relevant to airway inflammation in asthma.7,8 In sputum supernatant exosomes, PRG2, EPX, RNASE2 and CLC proteins were more abundant in EA patients, highlighting their importance in eosinophils functions.9 Their release is crucial because these proteins modulate the immune response and inflammation in SEA, influencing disease mechanisms and severity. Furthermore, the presence of proteins that are unique to each phenotype and those that differ in abundance between these asthma phenotypes could have a significant impact on clinical practice. These proteins could not only serve as diagnostic tools to distinguish between the different asthma phenotypes, but could also open the door to new therapeutic targets as they may be involved in biological processes related to the inflammatory immune response. For example, the exclusivity of the asthma-associated protein FLG in NEA patients (from whom serum exosomes were obtained) could, according to our results, be associated with reduced epithelial permeability and IL-33/TSLP expression in this asthma phenotype, as well as a reduced Th2 inflammatory response. In contrast, FLG deficiency is associated with the opposite effects in EA patients.7 Among the exosomal proteins common to the two sample types, only PSMB1 was recognised in the complete eosinophil proteome and may be involved in asthma pathogenesis.6 Our results, with PSMB1 unique to serum exosomes, and PSMB1 and PSMB3 more abundant in sputum supernatant exosomes from EA patients, are consistent with the findings of Liu et al. and suggest a possible adverse effect on the protective effect of glucocorticoids (GCs) in desensitising β-2 adrenergic receptors in these patients, who have a more severe phenotype and are often treated with GCs.10
Given the high number of proteins in sputum supernatant exosomes, a broad diversity of biological processes in which they were implicated was observed, spread over a wider range of categories compared to serum exosome proteins. Serum exosome proteins are mainly linked to biological processes related to the immune system, which is strongly associated with asthma.11 Sputum supernatant exosome proteins were associated with highly active processes (metabolism, cell cycle and signalling). In particular, the FcɛRI receptor signalling process, a crucial receptor in the context of asthma, especially in allergic asthma,12 was shared by both samples, indicating that the proteins present in the exosomes of two sample types play a prominent role in asthmatic disease at systemic and local levels.
In conclusion, the variability in the protein profile of exosomes purified from serum and sputum supernatant suggests that these different proteins, involved in multiple biological processes, highlight that EA and NEA phenotypes are very different, not only at the immunological level. This study, which represents a pioneering advance in the use of proteomics to differentiate between these two asthma phenotypes, highlights the potential of the proteome in the development of new diagnostic tools for disease. For example, it could allow the differentiation between these asthma phenotypes and the discovery of new therapeutic targets. The novel findings on exosomes in severe asthma are remarkable despite the small sample size. The main limitation is the lack of healthy controls or mild asthmatics, which are necessary to validate the observations.
Ethical ApprovalThe study protocol was approved by the Ethics Committees of the above-mentioned hospitals (Hospital Universitario Fundación Jiménez Díaz and Hospital Universitario de Navarra), and the study was conducted in accordance with the principles set forth in the Declaration of Helsinki.
FundingThis work was supported by ISCIII – Instituto de Salud Carlos III and co-funded by the European Union, FIS (Fondo de Investigación Sanitaria – Spanish Health Research Fund) grants PI21/00896 and FI19/00067; Miguel Servet Program (CP23/00017); CIBER de Enfermedades Respiratorias (CIBERES), ISCIII; and FEDER funds (Fondo Europeo de Desarrollo Regional).
Authors’ ContributionsVdP conceived the manuscript and designed the study. MG-M, JMR-M, JAC, and ZG-dC processed the samples, performed the experiments, and analysed the data. MG-M, JMR-M, JAC and VdP wrote and edited the manuscript. JS, MJR-N, SQ and JMO recruited the patients and obtained the samples and clinical data. All authors contributed and approved the submitted version.
Informed ConsentSigned informed consent was obtained from all patients.
Conflicts of InterestsJMR-M reports receiving payments for lectures and educational events form AstraZeneca and GSK. JS reports serving as a consultant to SANOFI, ABBIES and NOVARTIS; acquiring lecture fees by SANOFI, GSK, and Faes Farma; receiving support for attending meetings and/or travel by SANOFI; holding unpaid Leadership or fiduciary roles in other boards, societies, committees, or advocacy groups in SEAIC, AAAAI, and EAACI; and obtaining grant support for research from SANOFI. MJR-N reports receiving grant support for research from AstraZeneca and getting payments for lectures by AstraZeneca and SANOFI. SQ reports acquiring personal fees outside the submitted work from AstraZeneca, Novartis, SANOFI, Boehringer Ingelheim, Teva, ALK, Mundipharma, GSK, Chiesi and Leti. JMO reports obtaining grants from SANOFI during performance of the study and receiving personal fees outside of the submitted work from AstraZeneca and Mundipharma. VdP reports getting honoraria (advisory board, speaker) and/or institutional grant/research support from AstraZeneca and GSK and holding unpaid Leadership or fiduciary role in committee in EAACI. The rest of authors declare no conflicts of interest.
The authors would like to express their gratitude to all the patients for their voluntary participation, as well as all technical and nursing staff involved in the project (Manuela Garcia del Potro, Erica Aguado Wakui, Esther Gamella Álvarez, María Remedios Marquina Valero, Almudena Batanero Rodríguez, Raquel García Latorre and Zahara García de Castro). The authors also recognise the Proteomics Unit of the Universidad Complutense de Madrid (UCM), where the analysis of the described results was conducted. This work was supported by ISCIII – Instituto de Salud Carlos III and co-funded by the European Union, FIS (Fondo de Investigación Sanitaria – Spanish Health Research Fund) grants PI21/00896 and FI19/00067; Miguel Servet Program (CP23/00017); CIBER de Enfermedades Respiratorias (CIBERES), ISCIII; and FEDER funds (Fondo Europeo de Desarrollo Regional).