To investigate the microbiota and metabolome of patients with ABO compared with bronchiectasis and asthma, and determine the relevance with clinical characteristics, inflammatory endotype and exacerbation risks.
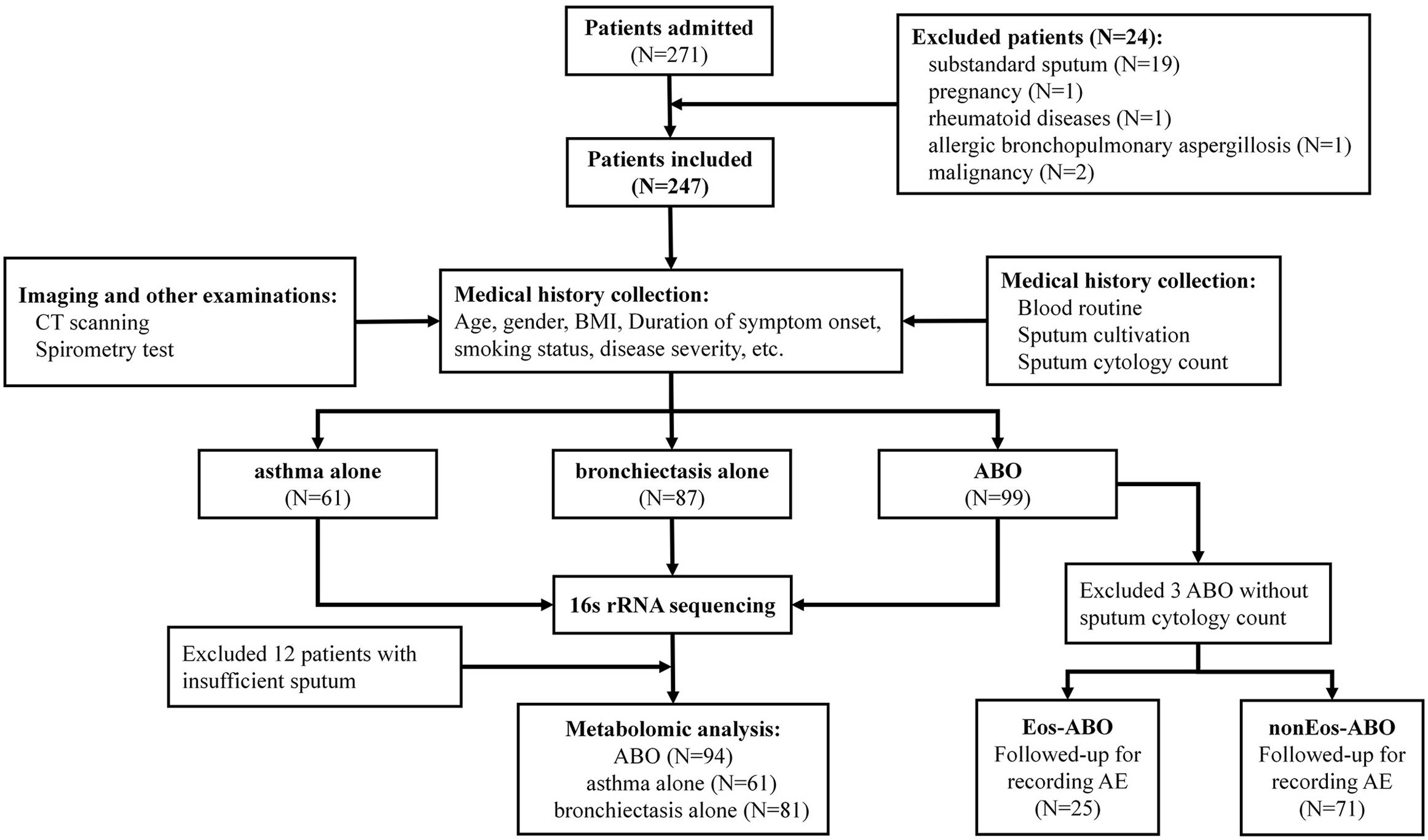
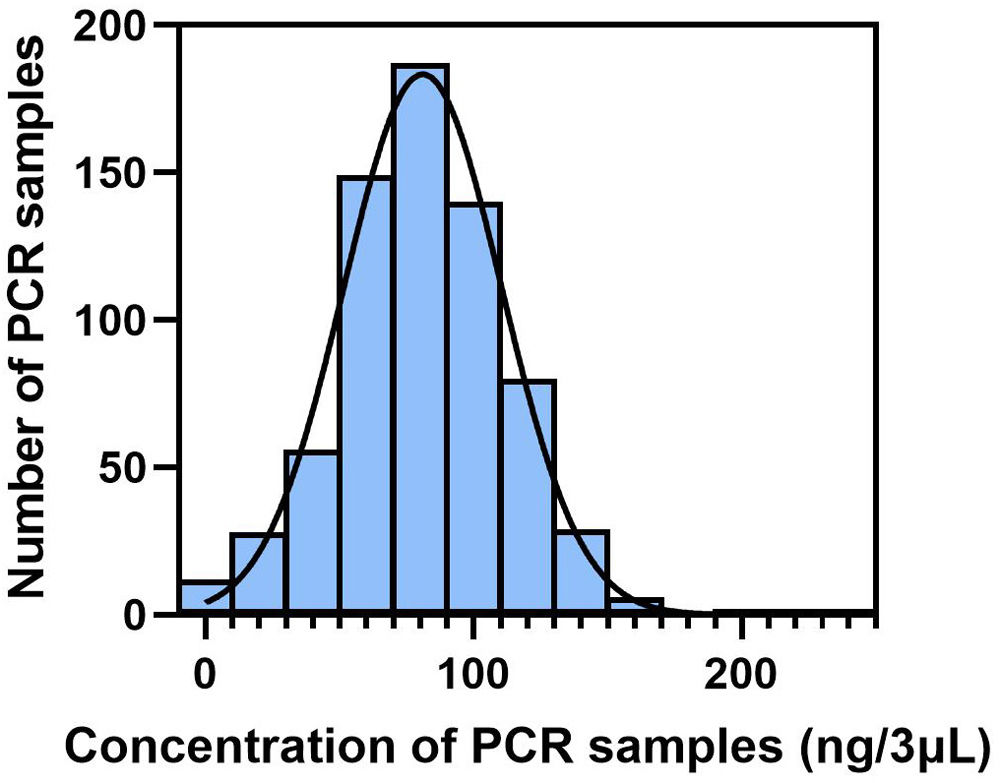
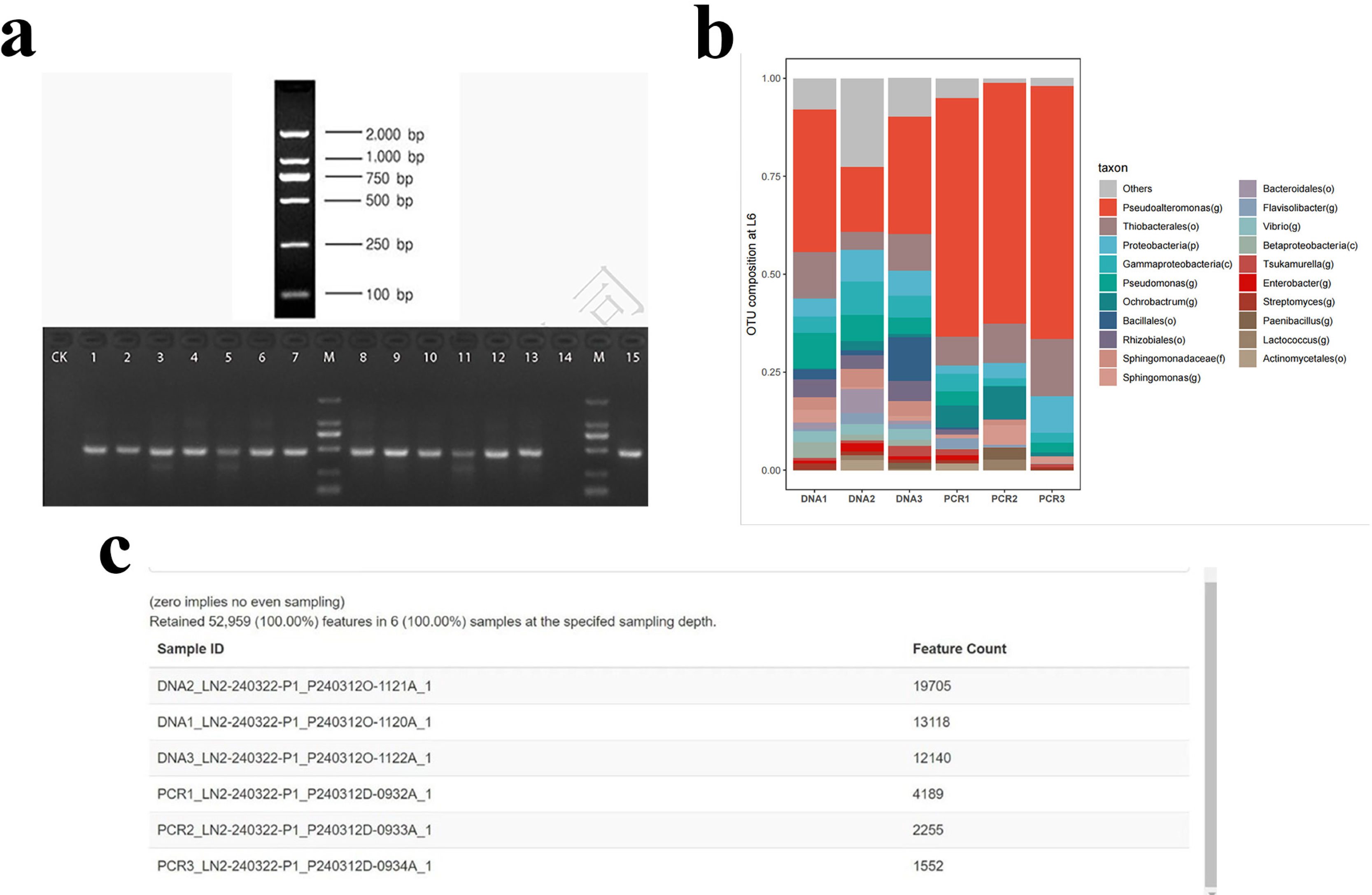
MethodsIn this prospective cohort study, patients underwent comprehensive assessments, including sputum differential cell count, and sputum collection at baseline. Sputum microbiota was profiled via 16S rRNA gene sequencing and metabolome via liquid chromatography/mass spectrometry. Shannon-Wiener Diversity Index (SWDI) was used to reflect dysbiosis. Patients were followed-up to record exacerbations. ABO patients were stratified by the SWDI and sputum eosinophilia to determine the exacerbation risks.
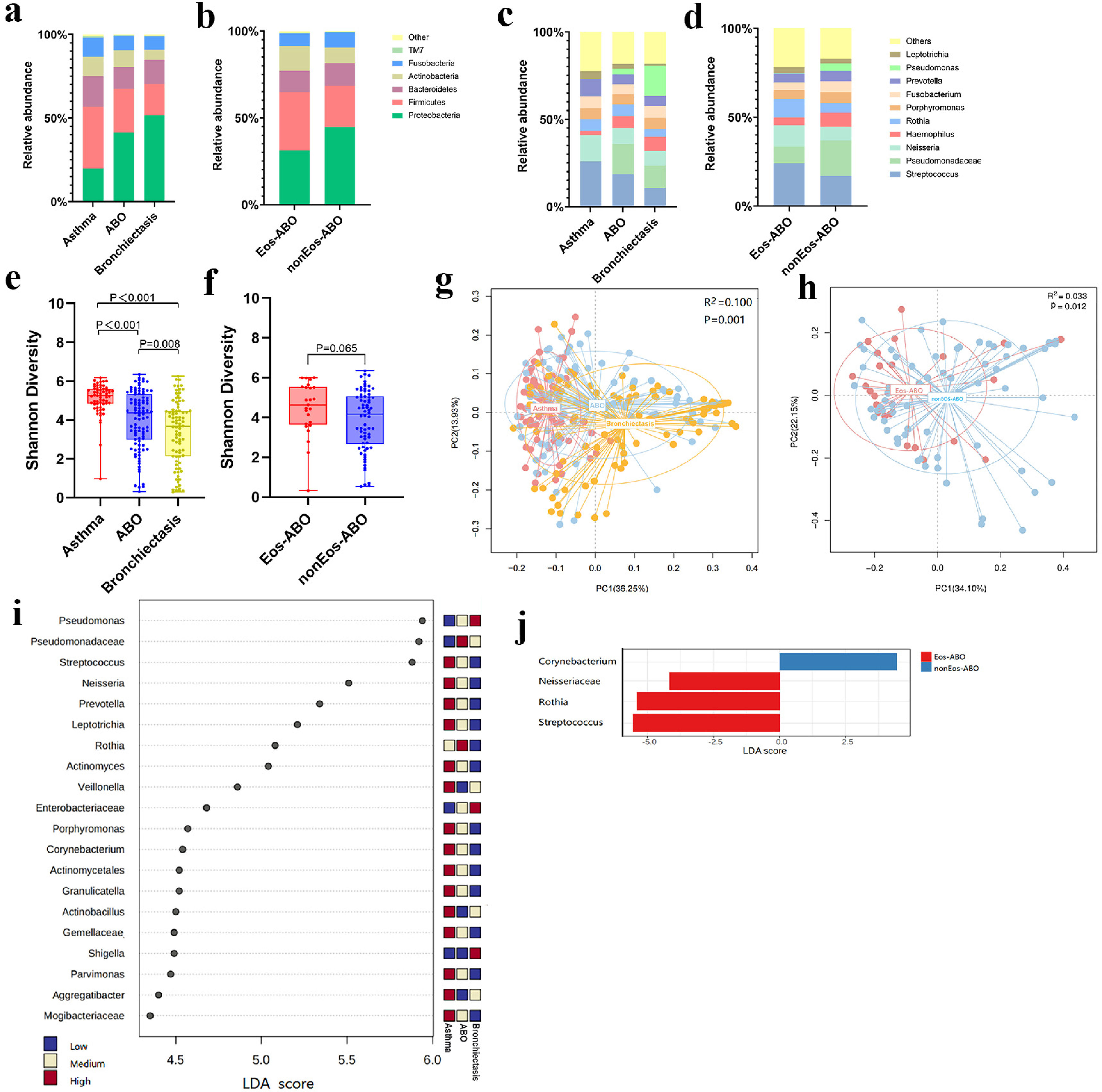
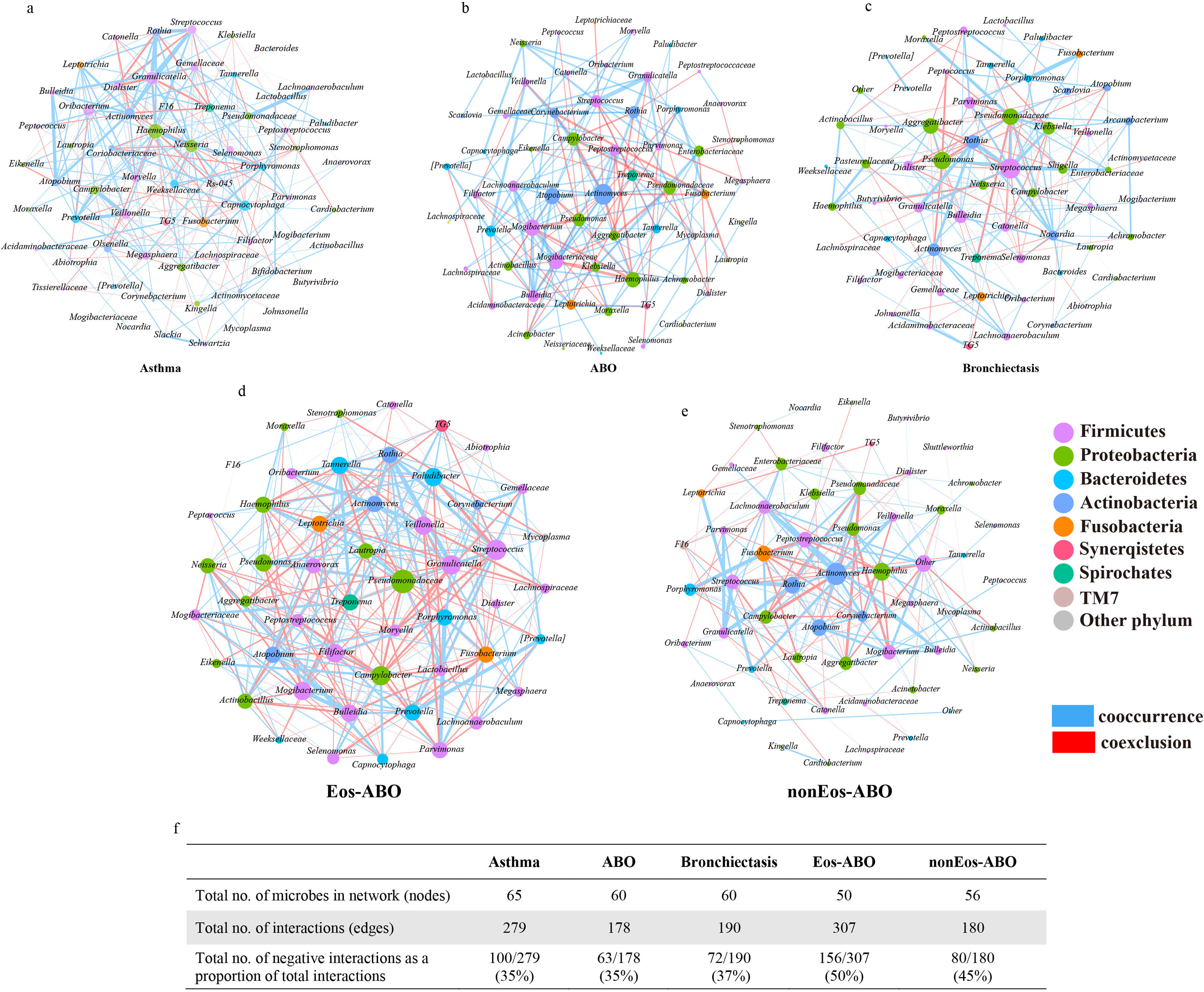
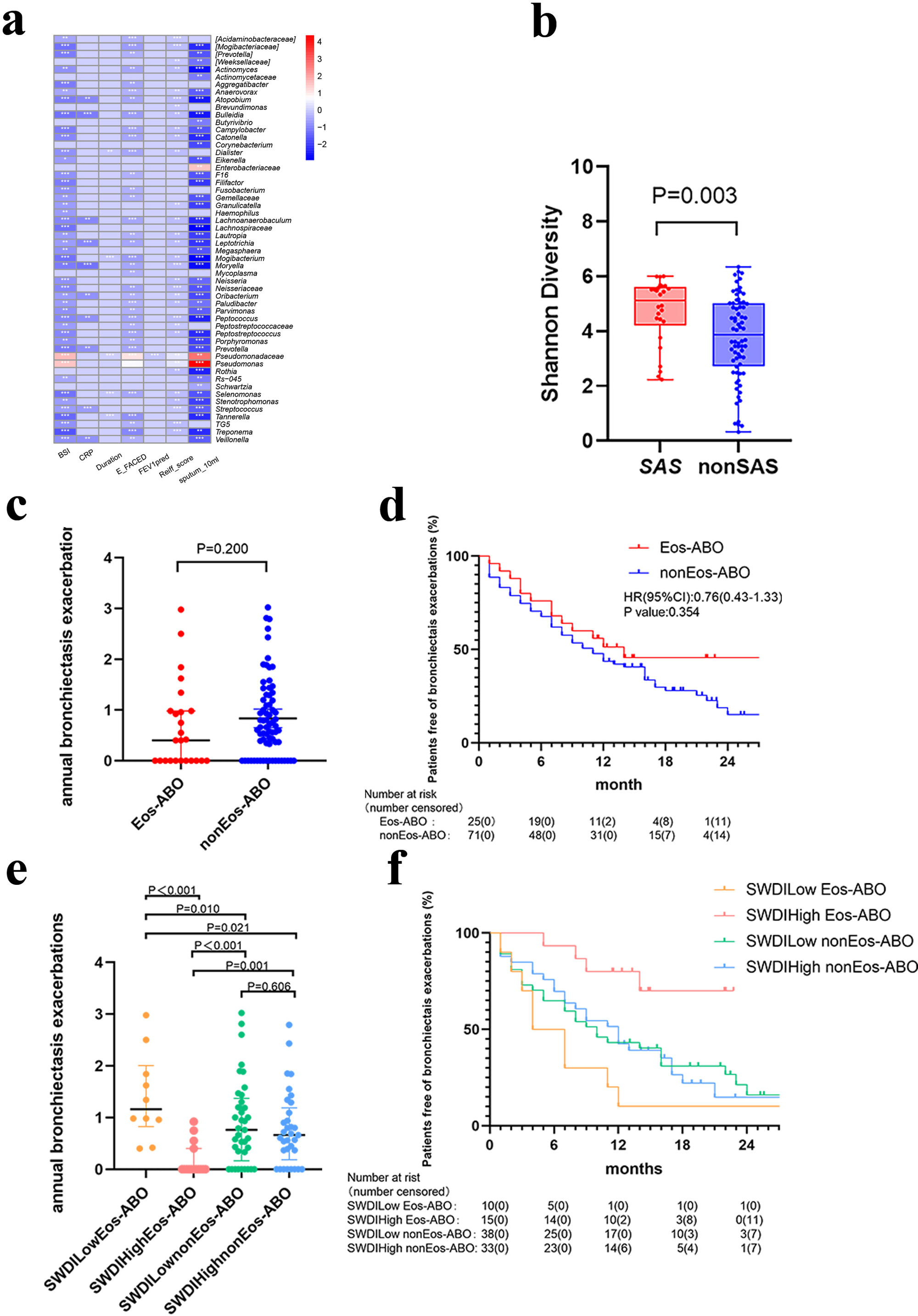
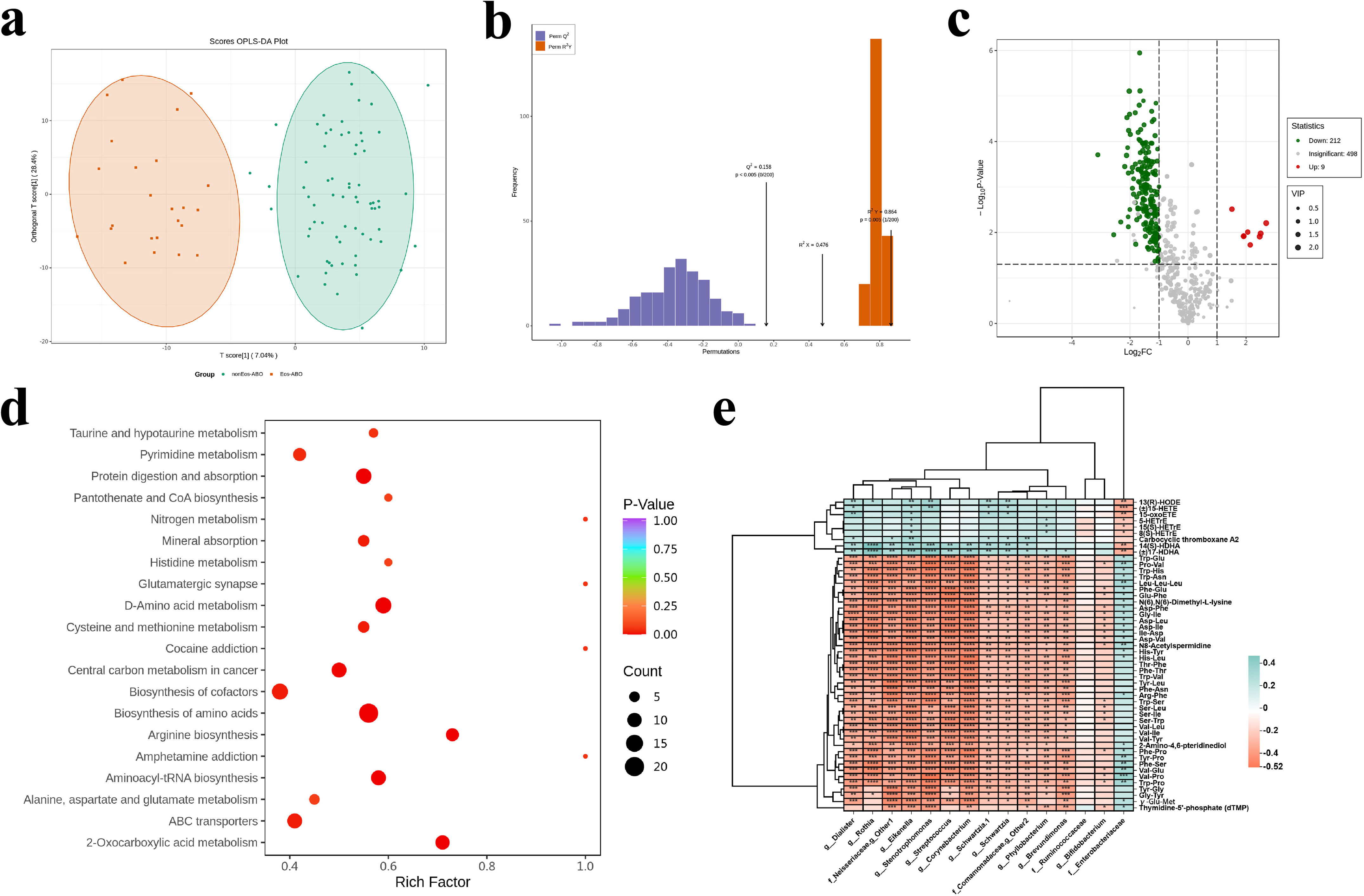
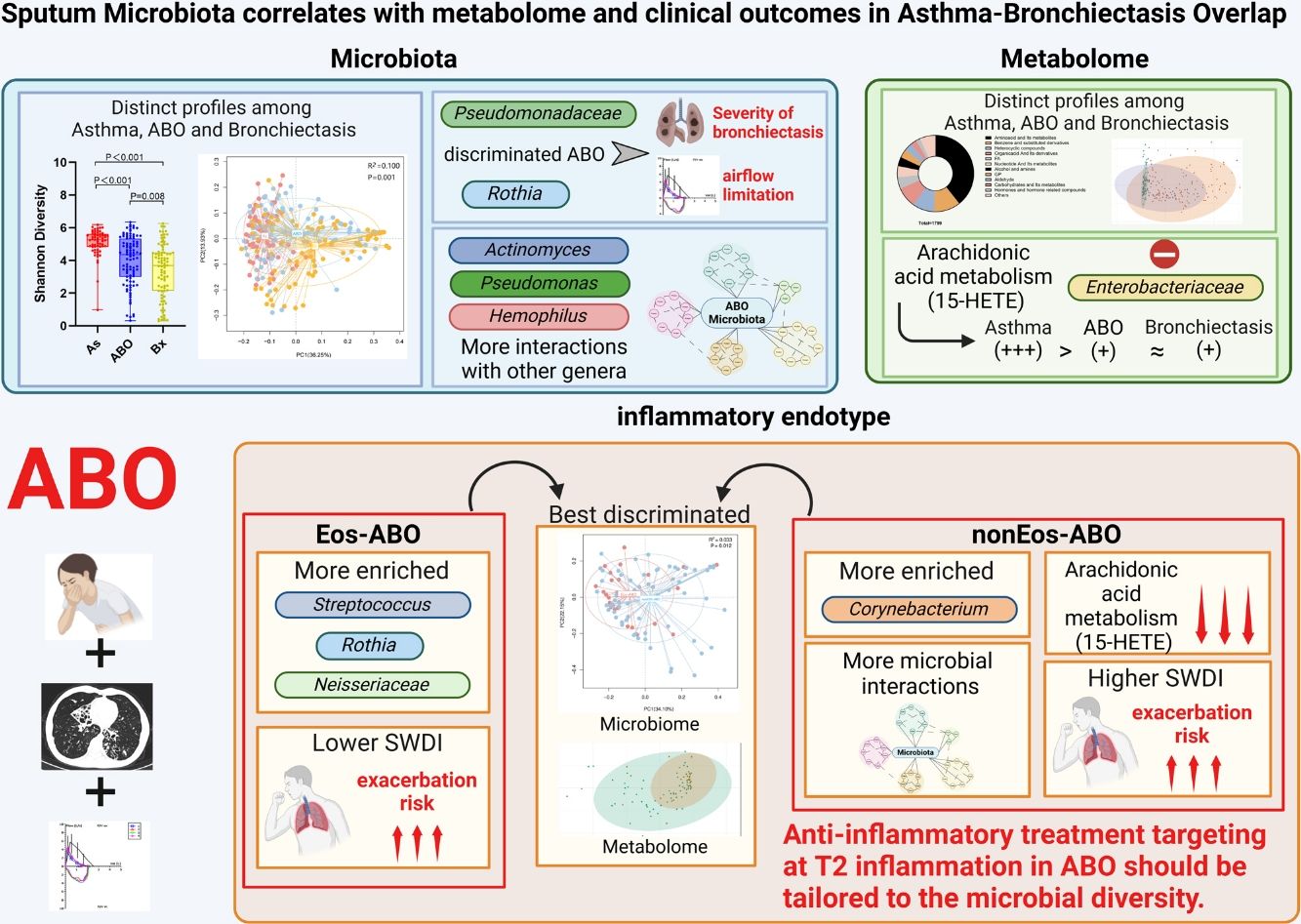
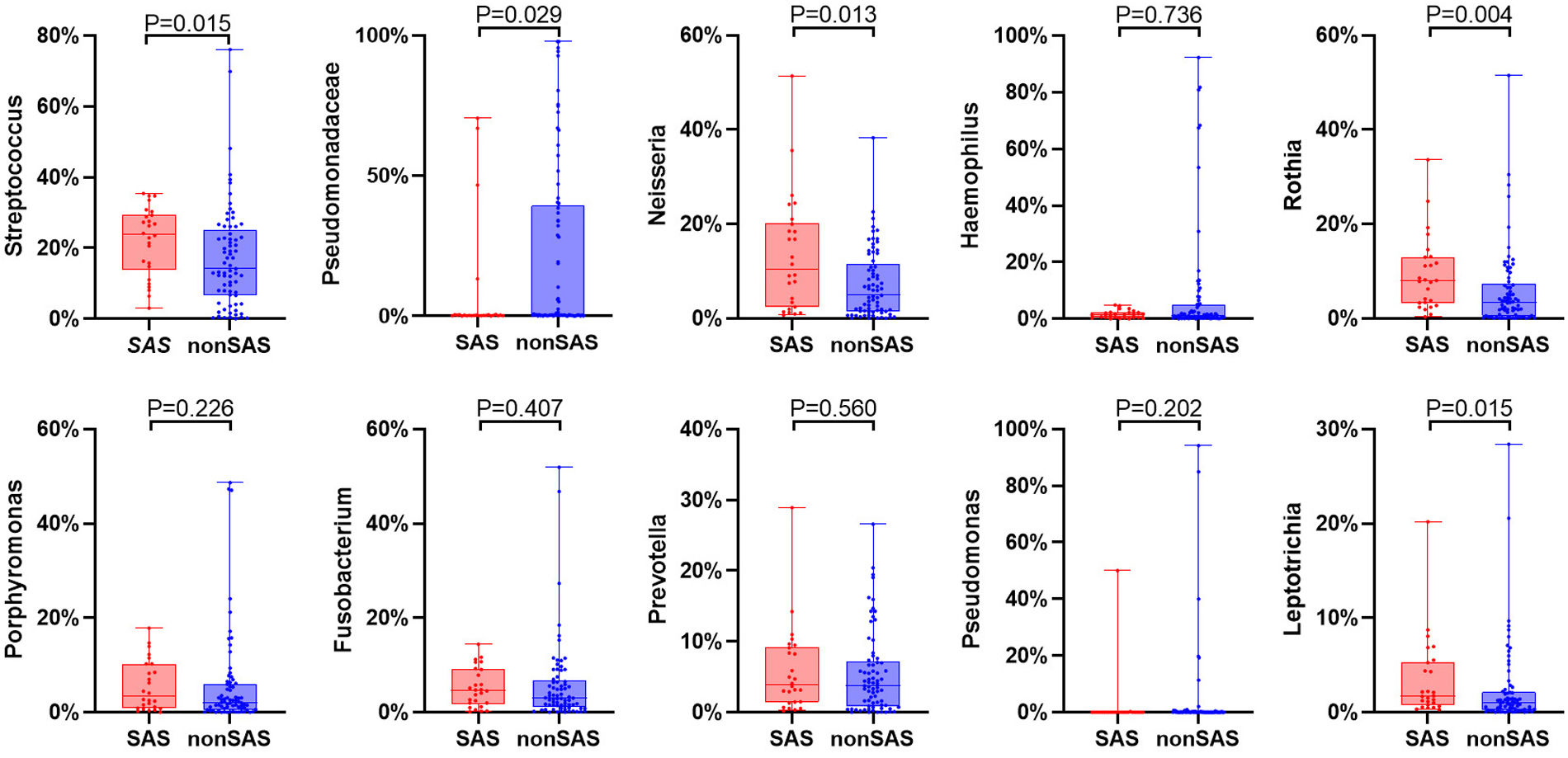
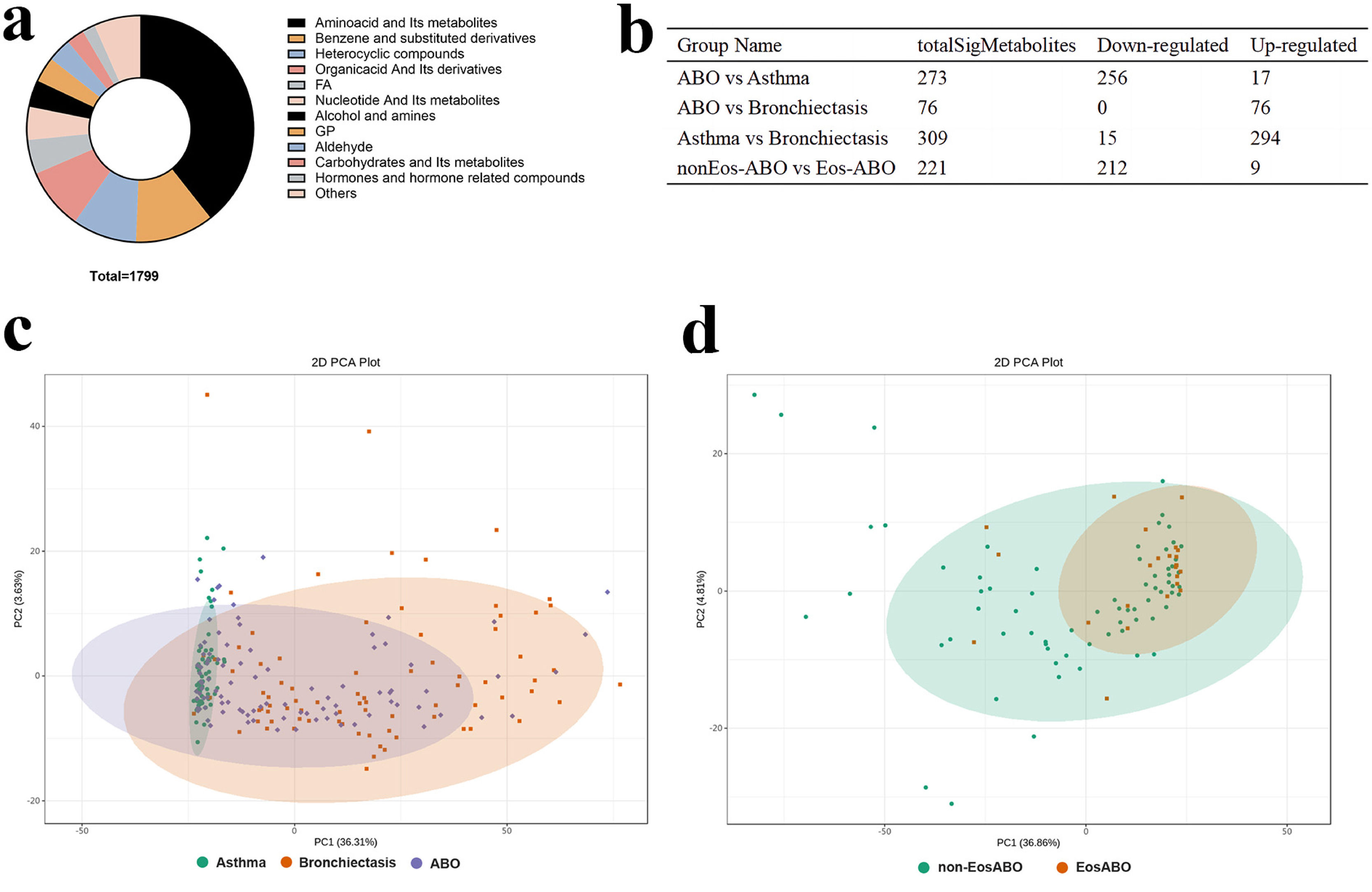
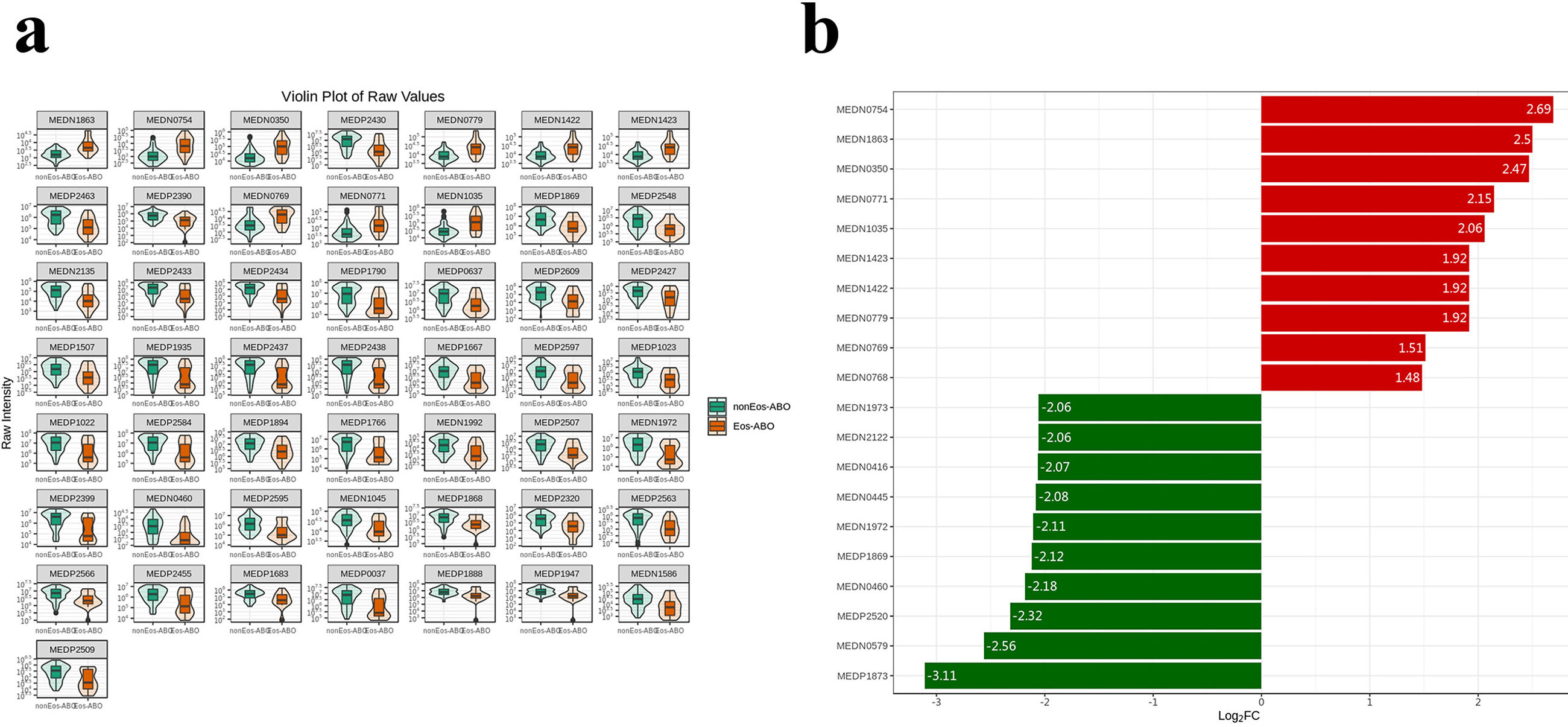
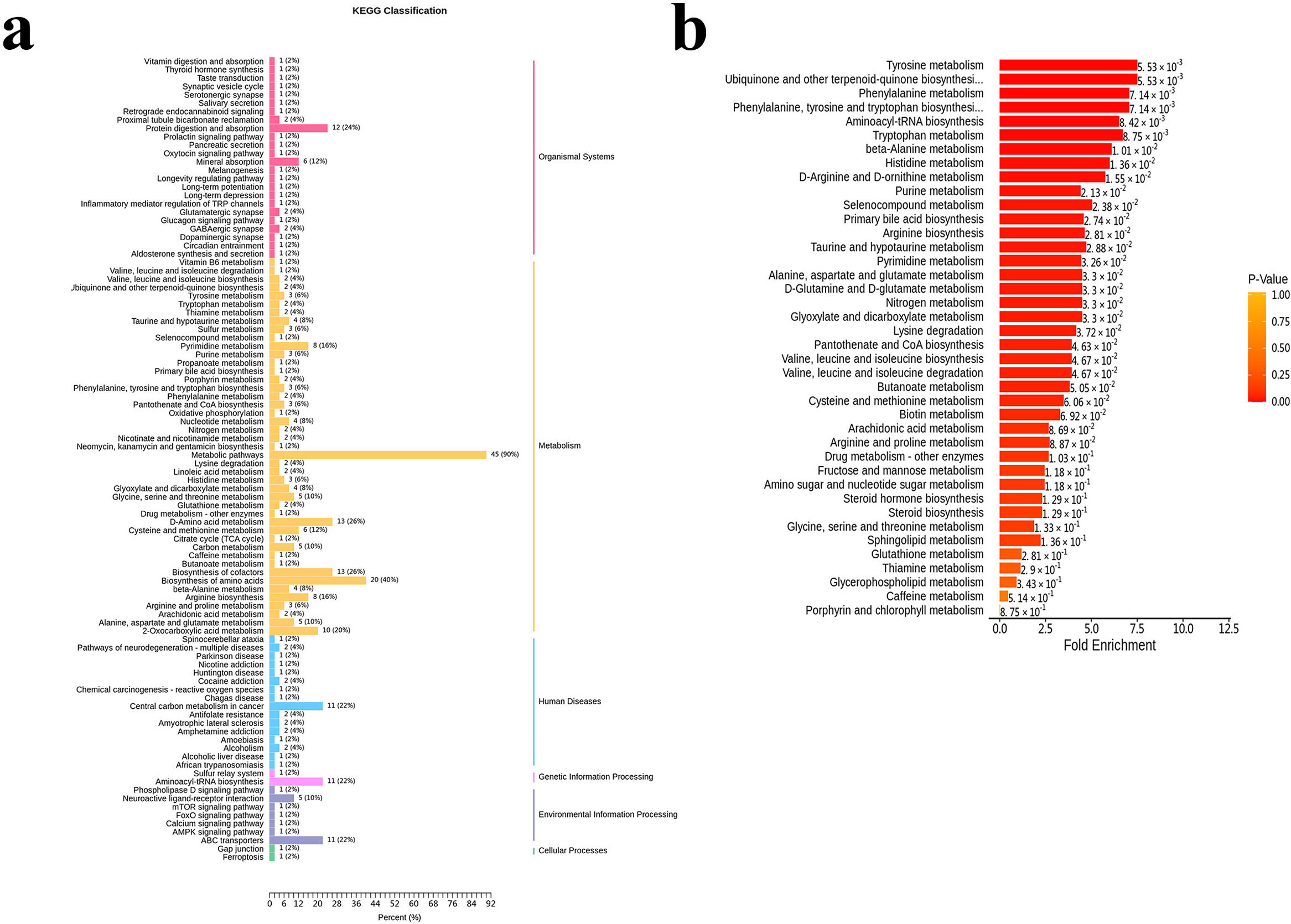
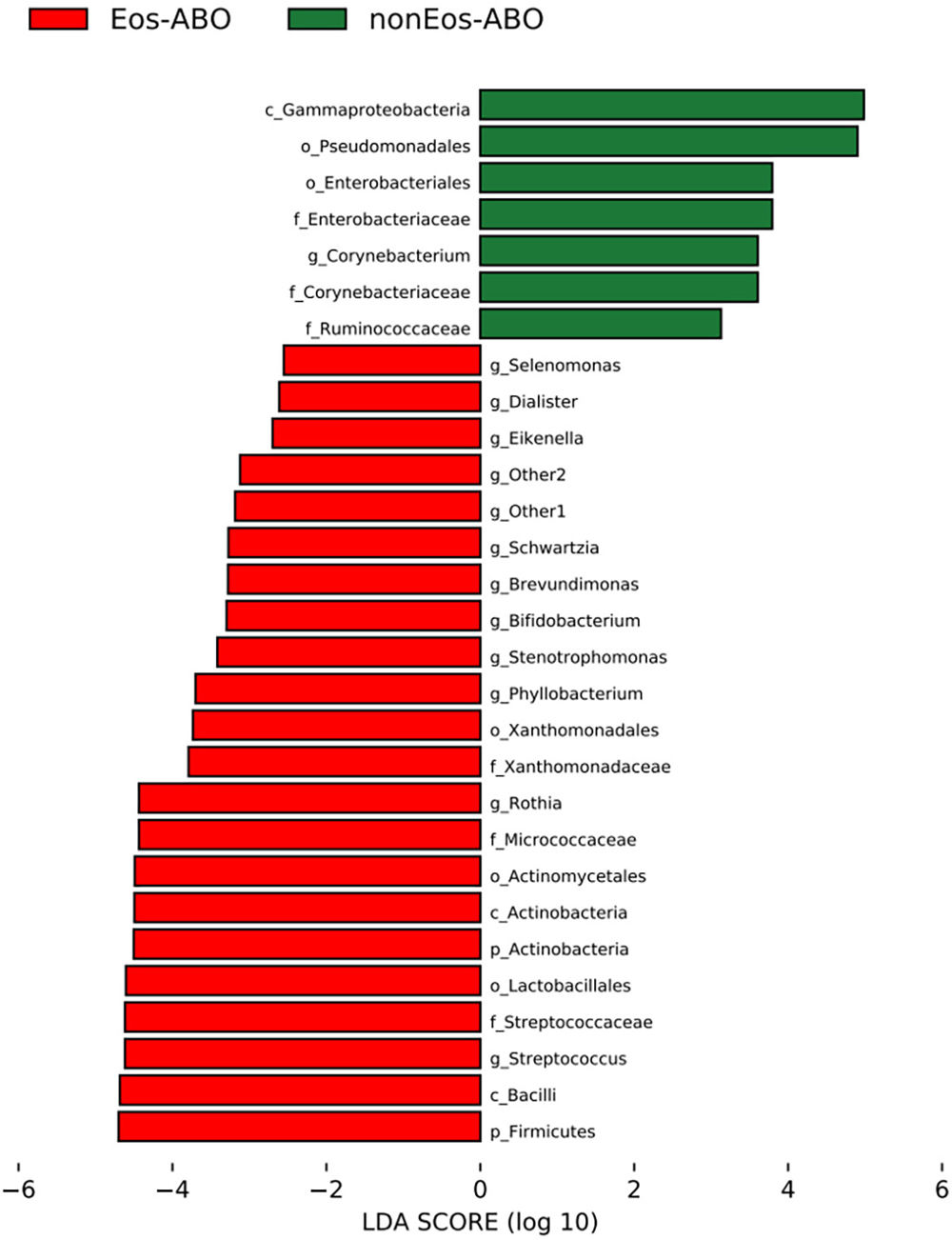
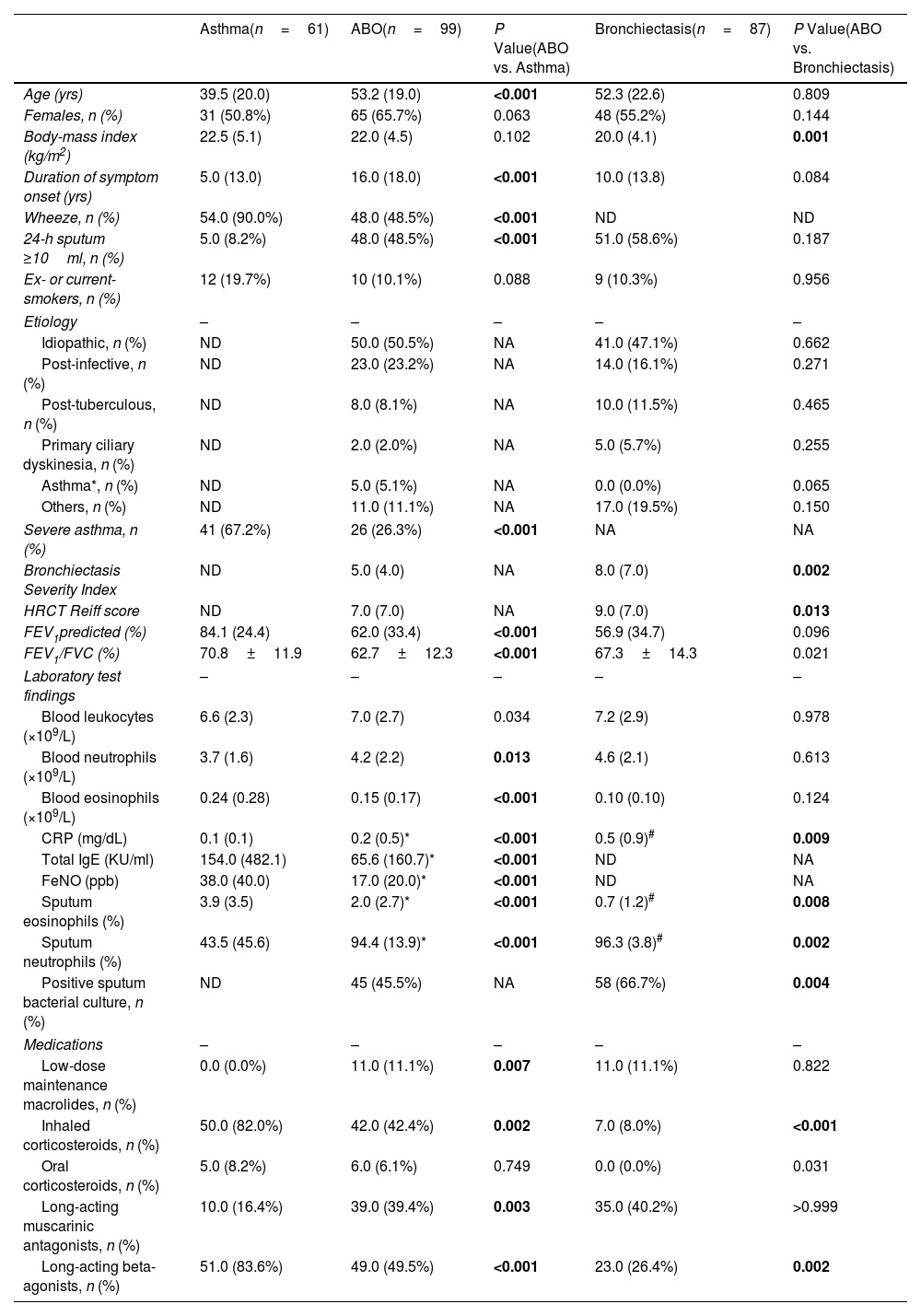
ResultsTwo hundred forty-seven patients were recruited, including 99 ABO (median age: 53.2 years, 65.7% female), 61 asthma (median age: 39.5 years, 50.8% female) and 87 bronchiectasis patients (median age: 52.3 years, 55.2% female). Both microbiota compositions and metabolites differed among asthma, ABO and bronchiectasis, and between eosinophilic and non-eosinophilic ABO at steady-state. Baseline SWDI of microbiota was highest in asthma, followed by ABO. Both Pseudomonadaceae and Rothia most effectively discriminated ABO from asthma and bronchiectasis. Pseudomonas exhibited a more pronounced negative correlation with other taxa in nonEos-ABO. ABO patients with low SWDI with sputum eosinophilia, or those with high SWDI without sputum eosinophilia, had a shorter time to the first exacerbation. Metabolomic compositions in Eos-ABO separated from nonEos-ABO. The relative abundance of Enterobacteriaceae correlated negatively with 15-hydroxylated eicosatetraenoic acid, whose concentrations were higher in Eos-ABO.
ConclusionsIntegrating microbiota and metabolome profiles, together with eosinophilic inflammatory endotyping, can inform exacerbation risk and personalized management of ABO.