No previous systematic reviews have comprehensively investigated the features of Xpert MTB/XDR and other rapid tests to diagnose pre-XDR/XDR-TB. The aim of this systematic review is to assess existing rapid diagnostics for pre-XDR/XDR-TB from a point-of-care perspective and describe their technical characteristics (i.e., sensitivity, specificity, positive and negative predictive values).
MethodsEmbase, PubMed, Scopus, and Web of Science were searched to detect the articles focused on the accuracy of commercially available rapid molecular diagnostic tests for XDR-TB according to PRISMA guidelines. The analysis compared the diagnostic techniques and approaches in terms of sensitivity, specificity, laboratory complexity, time to confirmed diagnosis.
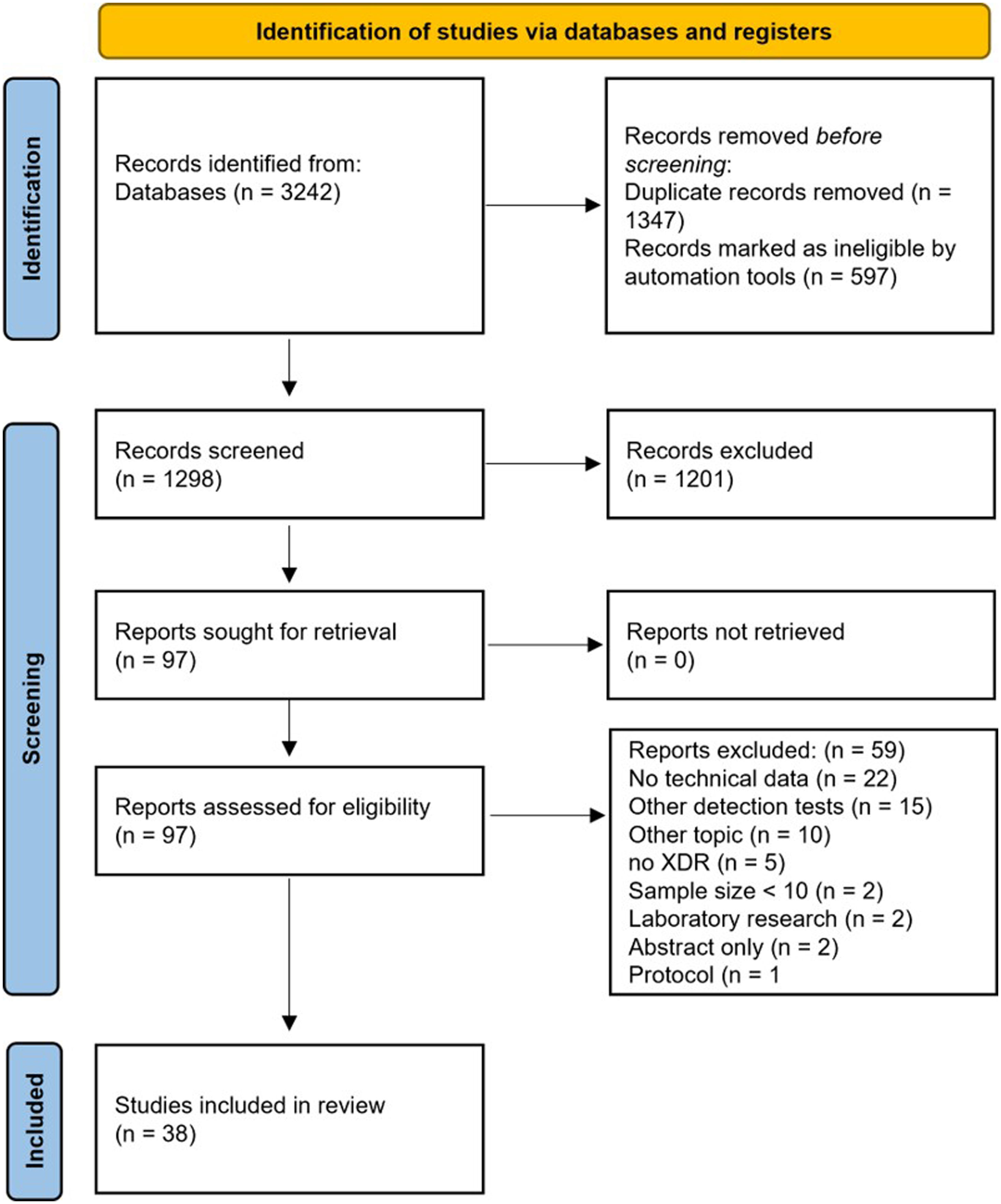
ResultsOf 1298 records identified, after valuating article titles and abstracts, 97 (7.5%) records underwent full-text evaluation and 38 records met the inclusion criteria.
Two rapid World Health Organization (WHO)-endorsed tests are available: Xpert MTB/XDR and GenoType MTBDRsl (VER1.0 and VER 2.0). Both tests had similar performance, slightly favouring Xpert, although only 2 studies were available (sensitivity 91.4–94; specificity 98.5–99; accuracy 97.2–97.7; PPV 88.9–99.1; NPV 95.8–98.9).
ConclusionsXpert MTB/XDR could be suggested at near-point-of-care settings to be used primarily as a follow-on test for laboratory-confirmed TB, complementing existing rapid tests detecting at least rifampicin-resistance.
Both Xpert MTB/XDR and GenoType MTBDRsl are presently diagnosing what WHO defined, in 2021, as pre-XDR-TB.
The appearance of drug-resistance has followed in the wake of extensive programmatic use of anti-tuberculosis (TB) drugs.1 The increasing problem of drug resistance poses different levels of diagnostic challenge: to identify the presence of drug resistance, define the extent of resistance present; and support the design of an effective anti-TB regimen.2
Defining the drug resistance profile and its severity. The appearance of strains of Mycobacterium tuberculosis resistant to both isoniazid and rifampicin, the two core anti-TB drugs, has defined multidrug-resistant tuberculosis (MDR-TB) from the outset. Later in 2006, when more resistant forms of TB appeared, the definition of extensively drug-resistant tuberculosis (XDR-TB) was agreed, referring to MDR-TB strains of M. tuberculosis with additional resistance to any fluoroquinolone and at least one of the three injectable anti-TB drugs (capreomycin, kanamycin and amikacin). The XDR-TB definition was made on the assumption that these classes of drugs were essential to successfully treat a patient with MDR-TB.3–5
More recent evidence demonstrated that XDR-TB patients had worse outcomes than MDR-TB patients with susceptibility to these drugs (those with resistance to fluoroquinolones faring worse than those resistant to injectables).6 The scientific community also considered codification of patterns of resistance beyond XDR7 and the possibility of defining so-called “total drug-resistance”, although no agreement was found on this.8,9
Although not formally recognized by the World Health Organization (WHO), clinicians widely adopted the term pre-XDR to indicate MDR-TB patients with additional resistance to either fluoroquinolones or injectables.9,10
In 2021 WHO introduced the definition pre-XDR-TB (e.g., MDR-TB plus resistance to fluoroquinolones) and modified the definition of XDR-TB, now being MDR-TB plus additional resistance to any fluoroquinolone and at least one WHO Group A drug (bedaquiline, linezolid).5,11,12
Diagnosing and treating pre- and XDR-TB. Considering the difficulty of treating XDR-TB, typically with costly and often toxic second-line anti-TB drugs, the importance of achieving rapid and effective diagnosis to design an effective regimen is obvious.
Of the 4.8 million people diagnosed with pulmonary TB worldwide in 2020, only 59% were bacteriologically confirmed and 71% of them were tested for rifampicin resistance. Notably, a WHO-recommended rapid molecular test was used as the initial diagnostic test for only 33% of new TB diagnoses, contributing to reasons why only about one third of the 157,903 MDR/rifampicin-resistant TB cases detected in 2020 had access to treatment.12
In a recent rapid communication,13,14 WHO recommended the use of shorter regimens on a programmatic basis. The pivotal initial tests which are used globally to classify patients as drug-susceptible or drug-resistant are the Xpert MTB/RIF and Xpert MTB/RIF Ultra, which allow detection of resistance to rifampicin only, and other new low-complexity molecular assays (Truenat MTB-RIF Dx) or moderate complexity nucleic acid amplification tests (NAATs) systems (Abbott Molecular RealTime MTB RIF/INH; Becton Dickinson BD MAX MDR-TB; Bruker-Hain Diagnostics FluoroType MTBDR; Roche Diagnostics Cobas MTB-RIF/INH) have been recently conditionally recommended as well.15 As the shorter MDR/RR-TB regimens are based on WHO Group A drugs, and no rapid WHO-approved tests are available for bedaquiline and linezolid, it is crucial that at least resistance to fluoroquinolones is rapidly detected, together with resistance to isoniazid.16 In case longer regimens are used, rapid detection of resistance to other drugs could be important (e.g., to injectables and ethionamide). Xpert MTB/XDR (isoniazid, ethionamide, fluoroquinolones, amikacin, capreomycin and kanamycin), first-line Line Probe Assays (LPAs; rifampicin and isoniazid) and second-line LPAs (fluoroquinolones, amikacin, capreomycin and kanamycin), plus other moderate complexity automated NAATs and high complexity reverse hybridization-based NAATs (Nipro Genoscholar PZA-TB) are the follow on rapid diagnostic tests currently recommended for the detection of additional drug-resistance beyond rifampicin.15,17
Despite advances, treatment success of MDR-TB remains low (globally at 59%); being lower for pre-XDR and XDR-TB patients and higher in settings where all oral regimens including new drugs are used.12,18,19 Improving treatment success will require access to rapid diagnostic tests able to reliably detect and classify drug resistance, and inform clinicians designing individual regimens.20
In 2021–2022 a Cochrane Review21,22 investigated the efficacy of Xpert MTB/XDR to rapidly detect resistance to isoniazid, fluoroquinolones, ethionamide, and amikacin (but not to the other injectable drugs kanamycin and capreomycin). However, no previous systematic reviews have comprehensively investigated the features of tests available to diagnose pre-XDR/XDR-TB. The aim of this systematic review is to assess all the tools able to rapidly diagnose pre-XDR/XDR-TB (including LPAs, and the Xpert XDR assay) under a point-of-care perspective and describe their technical characteristics (i.e., sensitivity, specificity, positive and negative predictive values).
MethodsSearch strategyFour electronic search engines repositories (i.e., Embase, PubMed, Scopus, and Web of Science) were searched for the detection of articles focused on the accuracy of rapid molecular diagnostic tests in the diagnosis of extensively drug-resistant tuberculosis (XDR-TB) cases.
The following keywords, combined in different strings, depending on the electronic database, were used to search articles published until February 2022: “Xpert MTB/XDR”, “drug susceptibility test”, “XDR-TB”, “sensitivity”, “specificity”, “tuberculosis”, “TB”, “Mycobacteriumtuberculosis”, “GenoType MTBDRsl”, and “line probe assay (LPA)”.
Reports published in the grey literature or in the social and conventional media were excluded following the risk of unreliable and poor scientific information on the adopted methodology. Reports that were not written in English language were excluded.
Study selectionOnly manuscripts describing the diagnostic accuracy of Xpert MTB/XDR compared to the drug susceptibility test (DST) were initially included.
All the studies with an experimental and observational design were selected with the exclusion of editorials, narrative reviews, case-reports or -series, laboratory studies, letters, study protocols, or correspondences.
Titles and abstracts were screened to evaluate the suitability of the manuscripts based on the above-mentioned inclusion and exclusion criteria.
The first assessment, carried out by two investigators (BDL and MVP), was supervised by a third investigator (GS). Full-texts were independently assessed by the same investigators and potential disagreement in the article selection process was resolved by consensus of the third investigator.
Study quality assessmentThis study was conducted and reported according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines.23
The Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) checklist was used to evaluate the methodological quality and applicability of the included studies.24 The following items were assessed: patient selection methods; index text description, conduction, and interpretation; reference standard (standard gold test) description, conduction, and interpretation.
Statistical analysisDescriptive statistics were used to summarize data. Meta-analytic estimates were computed and described with pooled and heterogeneity indicators. Forest plots were used to represent study variability with 95% confidence intervals (CI) for prevalence of drug resistance and for diagnostic accuracy of molecular techniques in the resistance identification. To assess the heterogeneity among studies the inconsistency indicator (I2) was calculated, where an I2 value>50% indicated substantial heterogeneity. Thus, fixed- and random-effects models were computed keeping into consideration the expected between-study heterogeneity. Bias assessment plots and Egger weighted regression test methods were used to assess the publication bias. A two-tailed p-value less than 0.05 was considered statistically significant. Data analyses were performed with the statistical softwares StatsDirect version 3.1.12 (StatsDirect Ltd.) and STATA version 17 (StatsCorp, College Station, Texas, USA).
ResultsStudy characteristicsThe search strategy identified 1298 records. After the evaluation of article titles and abstracts, 97 (7.5%) records underwent to the full-text evaluation. 3825–62 records met the inclusion criteria, whereas 59 (60.8%) were excluded for the following reasons: no technical data available (n=22), other detection tests (n=15), other topics (n=10), no XDR-TB isolates/samples included (n=5), sample size<10 (n=2), laboratory research (n=2), only abstract available (n=2), and only research protocol description (n=1) (Fig. 1).
Publication year ranged from 200962 to 202225 (Table 1). All the included studies had an observational design, and the minority were multicentric (8, 21.1%).25,28,30,32,42,43,47,48 The patient recruitment period ranged from 199254 to 2020,25 with a sample size ranging from 1753 to 264949 (Table 1). Following the list of WHO regions,63 20 reference centres enrolled patients from the European region,25,28,41,42,44,47–49,52,54,55,57,59,62 10 from the Western Pacific Region,28,32,34,36,38,45,46,51,60,61 8 from the African Region,25,30,39,40,43,48,56,58 6 from the South-East Asian Region,25,29,31,33,35,48,53 3 from the Eastern Mediterranean Region,26,27,37 3 from the Region of the Americas.28,50
Summary of the included studies.
| First author | Title | Year | Country | Mono-/multi-centre study | Study period | Sample size, n |
|---|---|---|---|---|---|---|
| Penn-Nicholson A | Detection of isoniazid, fluoroquinolone, ethionamide, amikacin, kanamycin, and capreomycin resistance by the Xpert MTB/XDR assay: a cross-sectional multicentre diagnostic accuracy study | 2022 | India, Moldova, South Africa | Multicenter | Jul 2019–Mar 2020 | 611 |
| Kardan-Yamchi J | Assessment of the GenoType MTBDRsl VER 2.0 compared to the phenotypic drug susceptibility testing and whole genome sequencing for the rapid detection of resistance to fluoroquinolone and second-line injectable drugs among rifampicin-resistant Mycobacterium tuberculosis isolates | 2021 | Iran | Monocenter | 35 | |
| Bouzouita I | Performance of the GenoType MTBDRsl V 2.0 for detecting second-line drugs resistance of Mycobacterium tuberculosis isolates in Tunisia | 2021 | Tunisia | Monocenter | May 2015–Dec 2019 | 103 |
| Cao Y | Xpert MTB/XDR: a 10-Colour Reflex Assay Suitable for Point-of-Care Settings To Detect Isoniazid, Fluoroquinolone, and Second-Line-Injectable-Drug Resistance Directly from Mycobacterium tuberculosis-Positive Sputum | 2021 | US, Georgia, Moldova, Peru, Vietnam, Italy | Multicenter | 314 | |
| Singh K | Direct detection of resistance to fluoroquinolones/SLIDs in sputum specimen by GenoType MTBDRsl v.2.0 assay A study from Eastern Uttar Pradesh, India | 2021 | India | Monocenter | Jan 2019–Dec 2019 | 225 |
| Ejo M | Effectiveness of GenoType MTBDRsl in excluding TB drug resistance in a clinical trial | 2021 | Africa | Multicenter | Jul 2012–Jun 2015 | 413 |
| Chandak RJ | Evaluation of MTBDRsl for detecting resistance in Mycobacterium tuberculosis to second-line drugs | 2019 | India | Monocenter | 205 | |
| Gao Y | Multi-centre evaluation of GenoType MTBDRsl line probe assay for rapid detection of pre-XDR and XDR Mycobacterium tuberculosis in China | 2018 | Province of China | Multicenter | 2015–2017 | 353 |
| Rufai SB | Association of gyrA and rrs gene mutations detected by MTBDRsl V1 on Mycobacterium tuberculosis strains of diverse genetic background from India | 2018 | India | Monocenter | 359 | |
| Jian J | Evaluation of the GenoType MTBDRplus and MTBDRsl for the detection of drug-resistant Mycobacterium tuberculosis on isolates from Beijing, China | 2018 | China | Monocenter | 2015–2016 | 96 |
| Yadav R | Diagnostic accuracy of GenoType(®) MTBDRsl VER 2.0 in detecting second-line drug resistance to M. tuberculosis | 2018 | India | Monocenter | 415 | |
| Zeng X | Performance of the MTBDRsl Line probe assay for rapid detection of resistance to second-line anti-tuberculosis drugs and ethambutol in China | 2017 | China | Monocenter | 162 | |
| Ennassiri W | Extensively drug-resistant tuberculosis (XDR-TB) in Morocco | 2017 | Morocco | Monocenter | 2015 | 155 |
| Lee YS | Performance of the GenoType MTBDRsl assay for the detection second-line anti-tuberculosis drug resistance | 2017 | South Korea | Monocenter | Dec 2011–Feb 2017 | 107 |
| Gardee Y | Evaluation of the GenoType MTBDRsl Version 2.0 Assay for Second-Line Drug Resistance Detection of Mycobacterium tuberculosis Isolates in South Africa | 2017 | South Africa | Monocenter | 2012–2014 | 268 |
| Maningi NE | Comparison of line probe assay to BACTEC MGIT 960 system for susceptibility testing of first and second-line anti-tuberculosis drugs in a referral laboratory in South Africa | 2017 | South Africa | Monocenter | Jan 2014–Jun 2014 | 97 |
| Tekin K | Evaluation of the BACTEC MGIT 960 SL DST Kit and the GenoType MTBDRsl Test for Detecting Extensively Drug-resistant Tuberculosis Cases | 2017 | Turkey | Monocenter | 46 | |
| Bang D | Performance of the GenoType MTBDRplus assay (v2.0) and a new extended GenoType MTBDRsl assay (v2.0) for the molecular detection of multi- and extensively drug-resistant Mycobacterium tuberculosis on isolates primarily from Lithuania | 2016 | Lithuania, Denmark | Multicenter | 2009–2010 | 43 |
| Tomasicchio M | The diagnostic accuracy of the MTBDRplus and MTBDRsl assays for drug-resistant TB detection when performed on sputum and culture isolates | 2016 | South Africa | Multicenter | 234 | |
| Brossier F | Performance of the New Version (v2.0) of the GenoType MTBDRsl Test for Detection of Resistance to Second-Line Drugs in Multidrug-Resistant Mycobacterium tuberculosis Complex Strains | 2016 | France | Monocenter | 2005–2015 | 127 |
| Lu W | Evaluation of MTBDRplus and MTBDRsl in Detecting Drug-Resistant Tuberculosis in a Chinese Population | 2016 | China | Monocenter | May 2008–Dec 2008 | 99 |
| Jeong HY | Evaluation of the GenoType® MTBDRsl assay in Korean patients with MDR or XDR tuberculosis | 2016 | Korea | Monocenter | Apr 2008–Jun 2008 | 40 |
| Tagliani E | Diagnostic Performance of the New Version (v2.0) of GenoType MTBDRsl Assay for Detection of Resistance to Fluoroquinolones and Second-Line Injectable Drugs: a Multicenter Study | 2015 | Italy, Sweden, Germany, Moldova | Multicenter | 2009–2013 | 459 |
| Catanzaro A | Performance Comparison of Three Rapid Tests for the Diagnosis of Drug-Resistant Tuberculosis | 2015 | India, Moldova, South Africa | Multicenter | 914 | |
| Simons SO | Molecular drug susceptibility testing in the Netherlands: performance of the MTBDRplus and MTBDRsl assays | 2015 | Netherlands | Monocenter | 2007–2012 | 2649 |
| Tukvadze N | Performance of the MTBDRsl assay in Georgia | 2014 | Georgia (USA) | Monocenter | Nov 2011–Apr 2012 | 138 |
| Jin J | Underestimation of the resistance of Mycobacterium tuberculosis to second-line drugs by the new GenoType MTBDRsl test | 2013 | China | Monocenter | 2006–2009 | 261 |
| Kontsevaya I | Diagnostic accuracy of the genotype MTBDRsl assay for rapid diagnosis of extensively drug-resistant tuberculosis in HIV-coinfected patients | 2013 | Russian | Monocenter | 2008–2010 | 90 |
| Singh AK | Rapid detection of drug resistance and mutational patterns of extensively drug-resistant strains by a novel GenoType® MTBDRsl assay | 2013 | India | Monocenter | Jan 2011–Aug 2012 | 17 |
| Lopez-Roa P | Susceptibility testing to second-line drugs and ethambutol by GenoType MTBDRsl and Bactec MGIT 960 comparing with agar proportion method | 2012 | Spain | Monocenter | 1992–2010 | 26 |
| Lacoma A | GenoType MTBDRsl for molecular detection of second-line-drug and ethambutol resistance in Mycobacterium tuberculosis strains and clinical samples | 2012 | Spain | Monocenter | 88 | |
| Barnard M | Genotype MTBDRsl line probe assay shortens time to diagnosis of extensively drug-resistant tuberculosis in a high-throughput diagnostic laboratory | 2012 | South Africa | Monocenter | Mar 2010–Sep 2011 | 516 |
| Miotto P | GenoType MTBDRsl performance on clinical samples with diverse genetic background | 2012 | Italy | Monocenter | 234 | |
| Said HM | Evaluation of the GenoType® MTBDRsl assay for susceptibility testing of second-line anti-tuberculosis drugs | 2012 | South Africa | Monocenter | 336 | |
| Zivanovic I | Detection of drug-resistant Mycobacterium tuberculosis strains isolated in Serbia by the genotype MTBDRsl assay | 2012 | Serbia | Monocenter | 2011 | 19 |
| Huang WL | Performance assessment of the GenoType MTBDRsl test and DNA sequencing for detection of second-line and ethambutol drug resistance among patients infected with multidrug-resistant Mycobacterium tuberculosis | 2011 | Taiwan | Monocenter | Jan 2008–Feb 2009 | 234 |
| Kiet VS | Evaluation of the MTBDRsl test for detection of second-line-drug resistance in Mycobacterium tuberculosis | 2010 | Vietnam | Monocenter | Jan 2005–Jul 2006 | 62 |
| Hillemann D | Feasibility of the GenoType MTBDRsl assay for fluoroquinolone, amikacin-capreomycin, and ethambutol resistance testing of Mycobacterium tuberculosis strains and clinical specimens | 2009 | Germany | Monocenter | 170 |
Among the included studies, two (5.3%),25,28 10 (26.3%),26,29,34,35,39,40,42,47,48,54 and 26 (68.4%)30–33,35–38,41,43–46,49–57,59–62 articles described the application of Xpert MTB/XDR, GenoType MTBDRsl VER 2.0, and GenoType MTBDRsl VER 1.0, respectively, for the detection of XDR-TB in strains or clinical isolates in comparison with the DST. The definition of XDR-TB used in the evaluated studies was the 2006 one.64
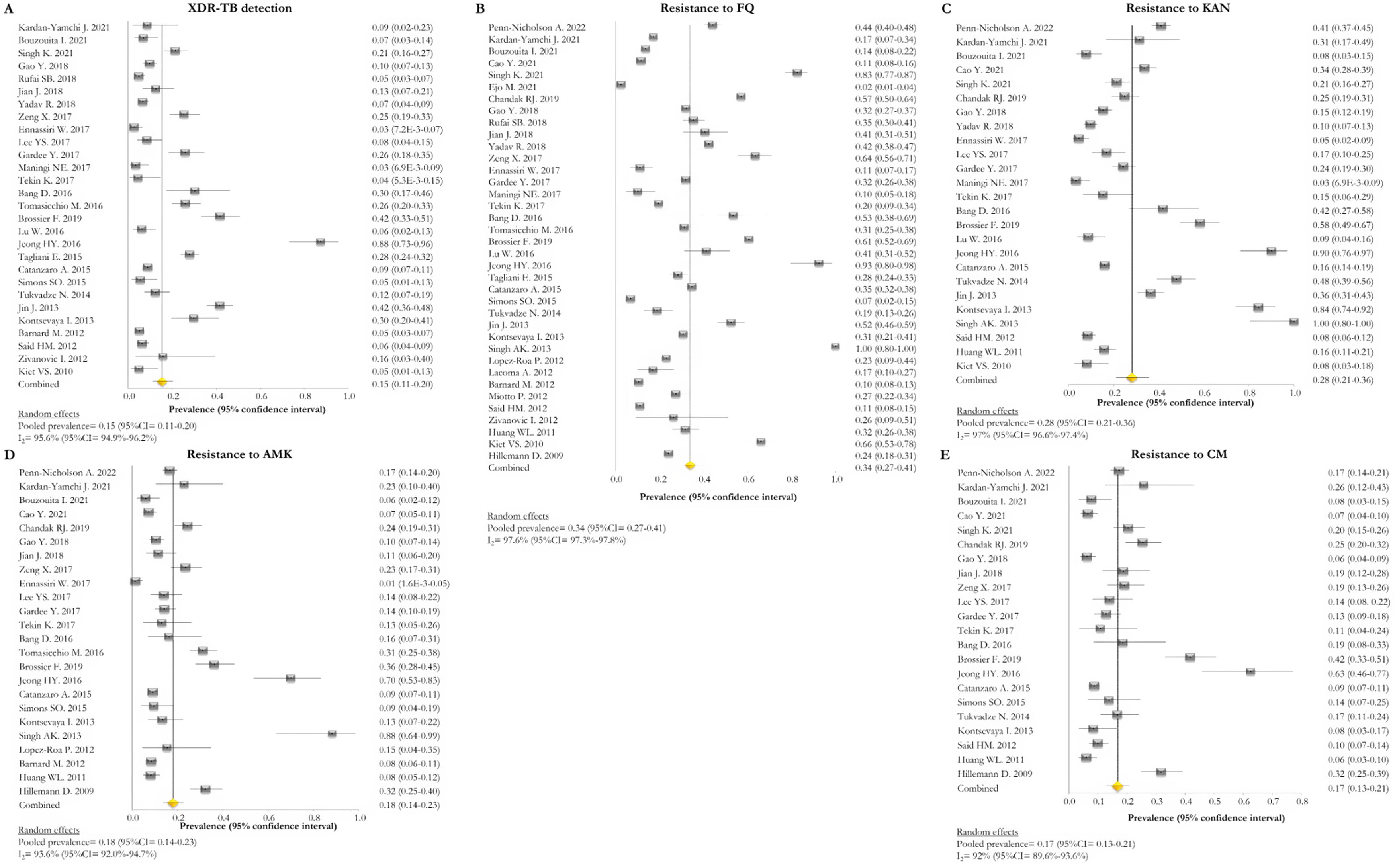
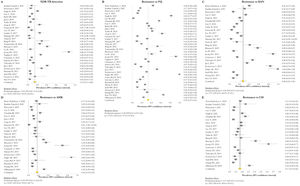
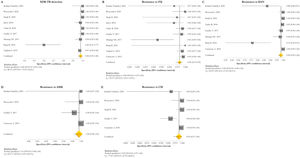
Resistance patternsThe overall pooled prevalence of XDR-TB pattern estimated by the DST was assessed in 28 (73.7%) studies26,27,29,32–52,56,58,59,61 and was 15% (95% CI=11–20; I2=95.6%). The pooled prevalence of resistance was higher for fluoroquinolones (FQ) (34%; 95% CI=27–41; I2=97.6%), followed by kanamycin (KAN) (28%; 95% CI=21–36; I2=97%), amikacin (AMK) (18%; 95% CI=14–23; I2=93.6%), and capreomycin (CM) (17%; 95% CI=13–21; I2=92%) (Fig. 2).
Forest plots of pooled prevalence of XDR-TB detection (A), and for resistance to FQ (B), KAN (C), AMK (D), and CM (E). The point estimates of prevalence from each study are indicated as a square and a 95% confidence interval is shown with a horizontal line; the yellow diamond is representative for the combined prevalence.
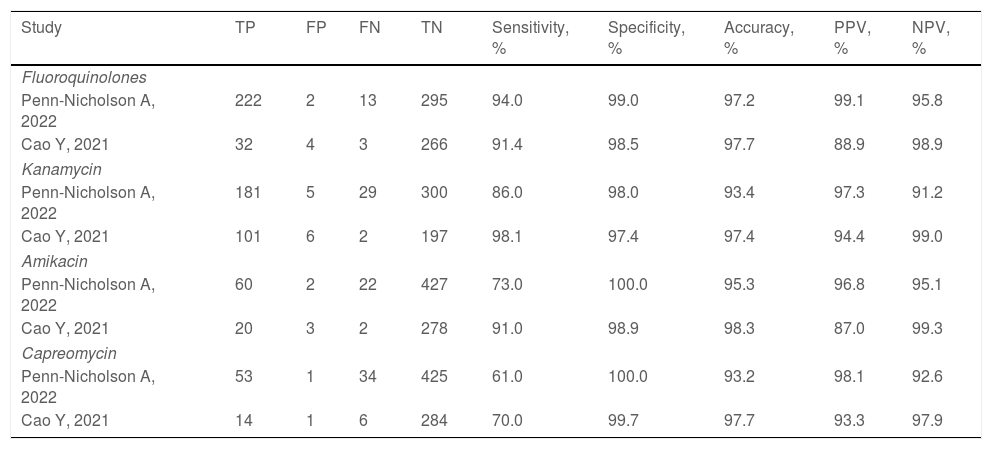
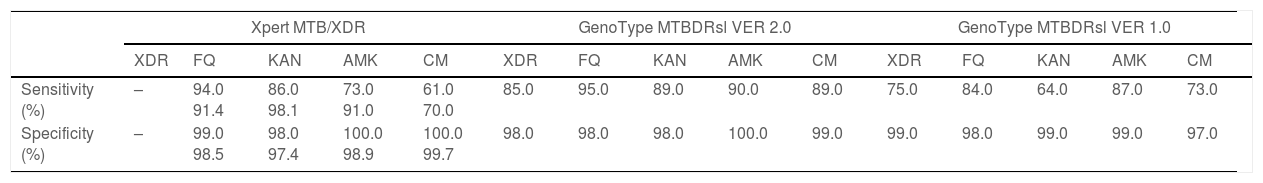
Two studies evaluated the diagnostic accuracy of the Xpert MTB/XDR test.25,28 Both assessed the resistance genes to FQ, KAN, AMK, and CM. Cao and colleagues estimated a sensitivity of 91.4%, 98.1%, 91.0%, and 70.0% for FQ, KAN, AMK, CM, respectively, whereas Penn-Nicholson and colleagues assessed a sensitivity of 94.0%, 86.0%, 73.0%, and 61.0% for FQ, KAN, AMK, CM (Table 2). Specificity and the other diagnostic characteristics for Xpert MTB/XDR were included in Table 2.
Diagnostic performance indicators of the Xpert MTB/XDR test.
| Study | TP | FP | FN | TN | Sensitivity, % | Specificity, % | Accuracy, % | PPV, % | NPV, % |
|---|---|---|---|---|---|---|---|---|---|
| Fluoroquinolones | |||||||||
| Penn-Nicholson A, 2022 | 222 | 2 | 13 | 295 | 94.0 | 99.0 | 97.2 | 99.1 | 95.8 |
| Cao Y, 2021 | 32 | 4 | 3 | 266 | 91.4 | 98.5 | 97.7 | 88.9 | 98.9 |
| Kanamycin | |||||||||
| Penn-Nicholson A, 2022 | 181 | 5 | 29 | 300 | 86.0 | 98.0 | 93.4 | 97.3 | 91.2 |
| Cao Y, 2021 | 101 | 6 | 2 | 197 | 98.1 | 97.4 | 97.4 | 94.4 | 99.0 |
| Amikacin | |||||||||
| Penn-Nicholson A, 2022 | 60 | 2 | 22 | 427 | 73.0 | 100.0 | 95.3 | 96.8 | 95.1 |
| Cao Y, 2021 | 20 | 3 | 2 | 278 | 91.0 | 98.9 | 98.3 | 87.0 | 99.3 |
| Capreomycin | |||||||||
| Penn-Nicholson A, 2022 | 53 | 1 | 34 | 425 | 61.0 | 100.0 | 93.2 | 98.1 | 92.6 |
| Cao Y, 2021 | 14 | 1 | 6 | 284 | 70.0 | 99.7 | 97.7 | 93.3 | 97.9 |
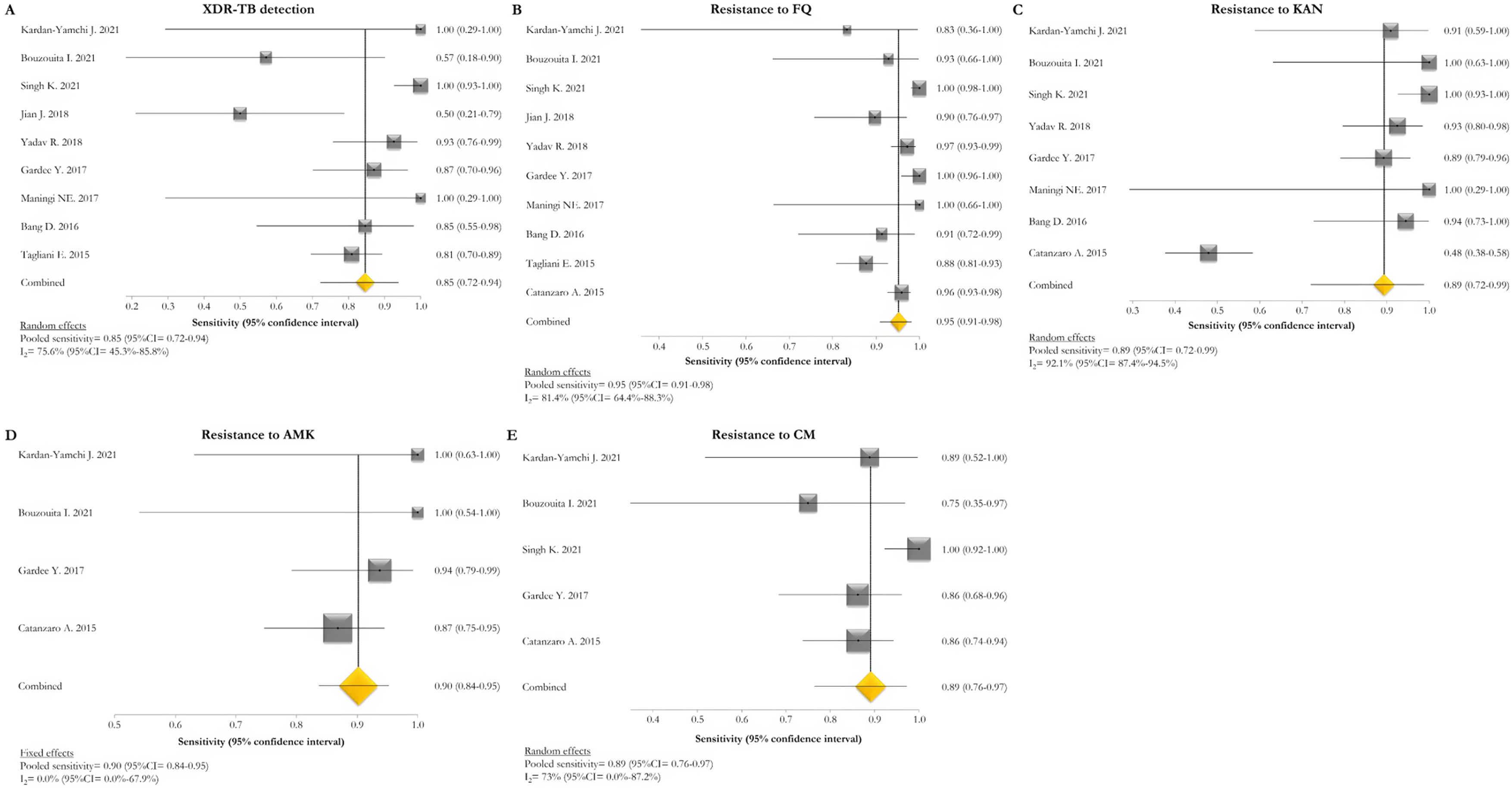
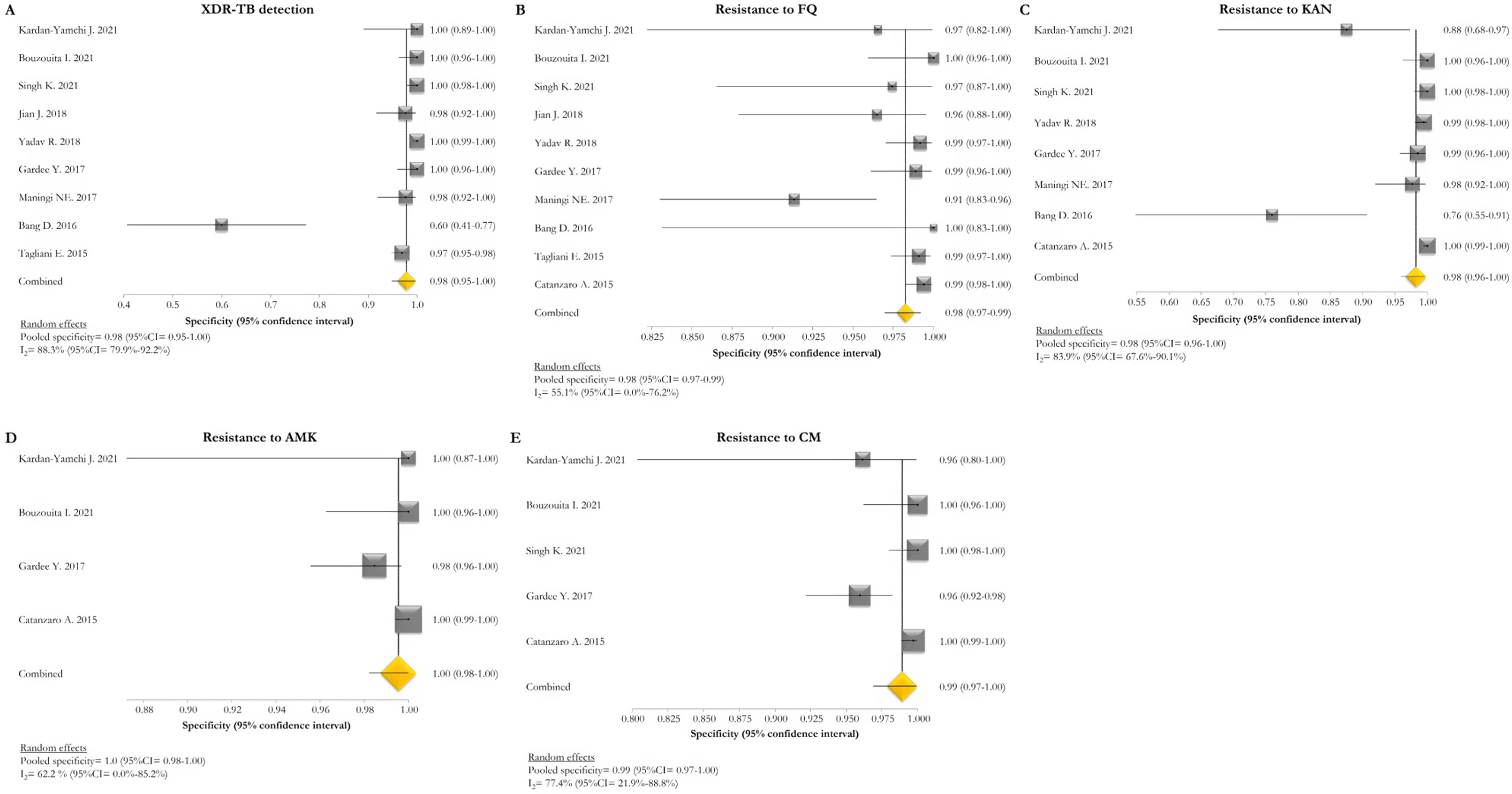
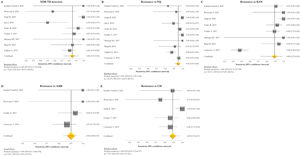
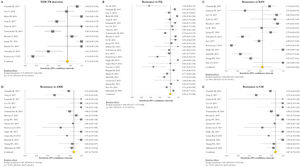
The pooled proportion of diagnostic sensitivity and specificity of GenoType MTBDRsl VER 2.0 in detecting XDR-TB were evaluated by 9 out of 10 (90%) studies,26,27,29,34,35,39,40,42,47 and were 85% (95% CI=72–94; I2=75.6%) and 98% (95% CI=95–100; I2=88.3%), respectively (Figs. 3 and 4).
Forest plots of pooled sensitivity of XDR-TB detection (A), and for resistance to FQ (B), KAN (C), AMK (D), and CM (E) of GenoType MTBDRsl VER 2.0. The point estimates of sensitivity from each study are indicated as a square and a 95% confidence interval is shown with a horizontal line; the yellow diamond is representative for the combined sensitivity.
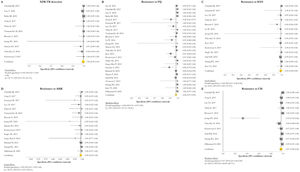
Forest plots of pooled specificity of XDR-TB detection (A), and for resistance to FQ (B), KAN (C), AMK (D), and CM (E) of GenoType MTBDRsl VER 2.0. The point estimates of specificity from each study are indicated as a square and a 95% confidence interval is shown with a horizontal line; the yellow diamond is representative for the combined specificity.
The pooled sensitivity and specificity for FQ resistance were high, reaching 95% (95% CI=91–98; I2=81.4%) and 98% (95% CI=97–99; I2=55.1%). When the test was used for the detection of second line injectable drugs (SLIDs), the pooled sensitivity and specificity estimates were: 89% (95% CI=72–99; I2=92.1%) and 98% (95% CI=96–100; I2=83.9%) for KAN; 90% (95% CI=84–95; I2=0%) and 100% (95% CI=98–100; I2=62.2%) for AMK; and 89% (95% CI=76–97; I2=73%) and 99% (95% CI=97–100; I2=77.4%) for CM. The diagnostic variables, including accuracy, PPV, and NPV, for XDR-TB detection and for resistance to FQ, KAN, AMK, and CM were described in Table S1.
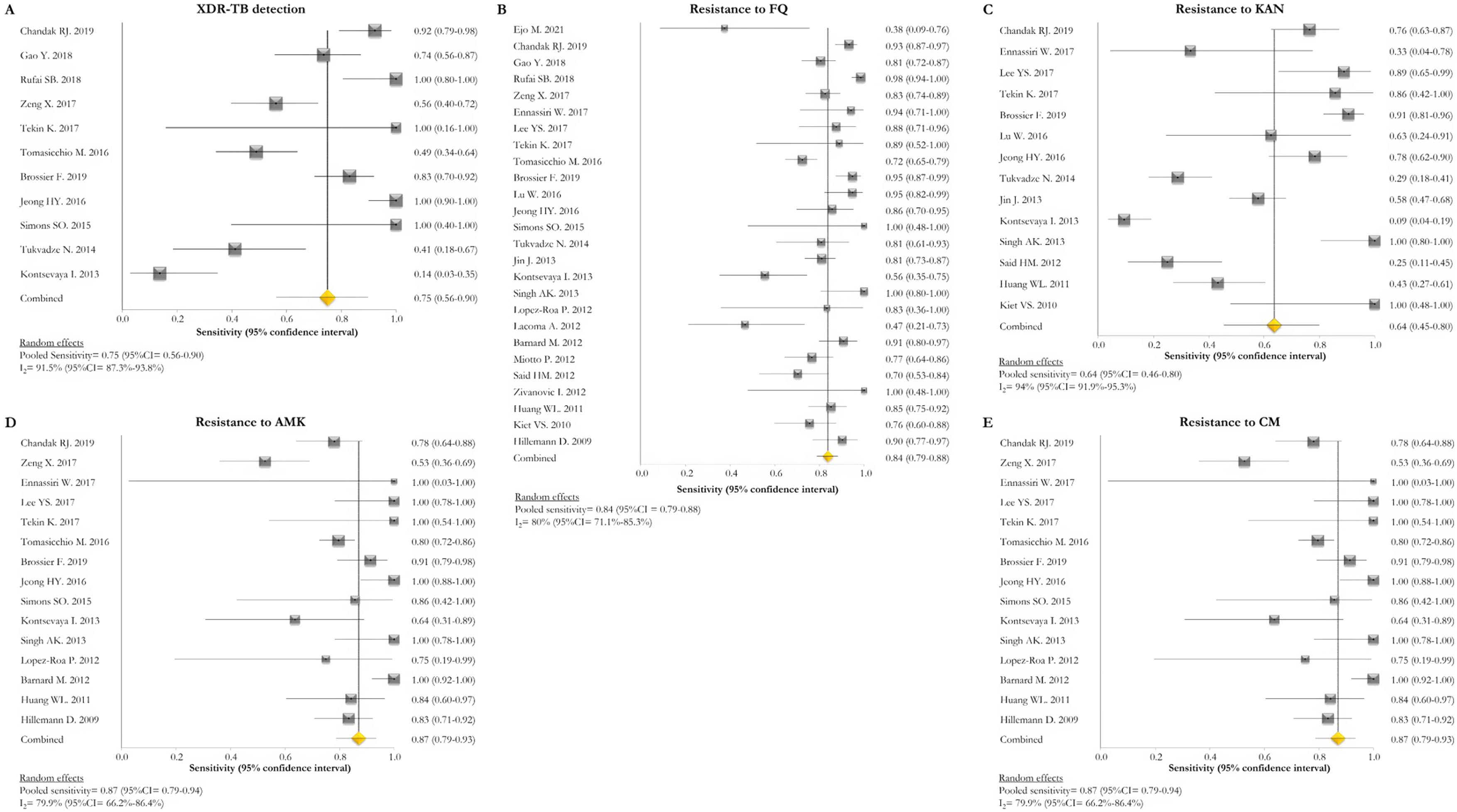
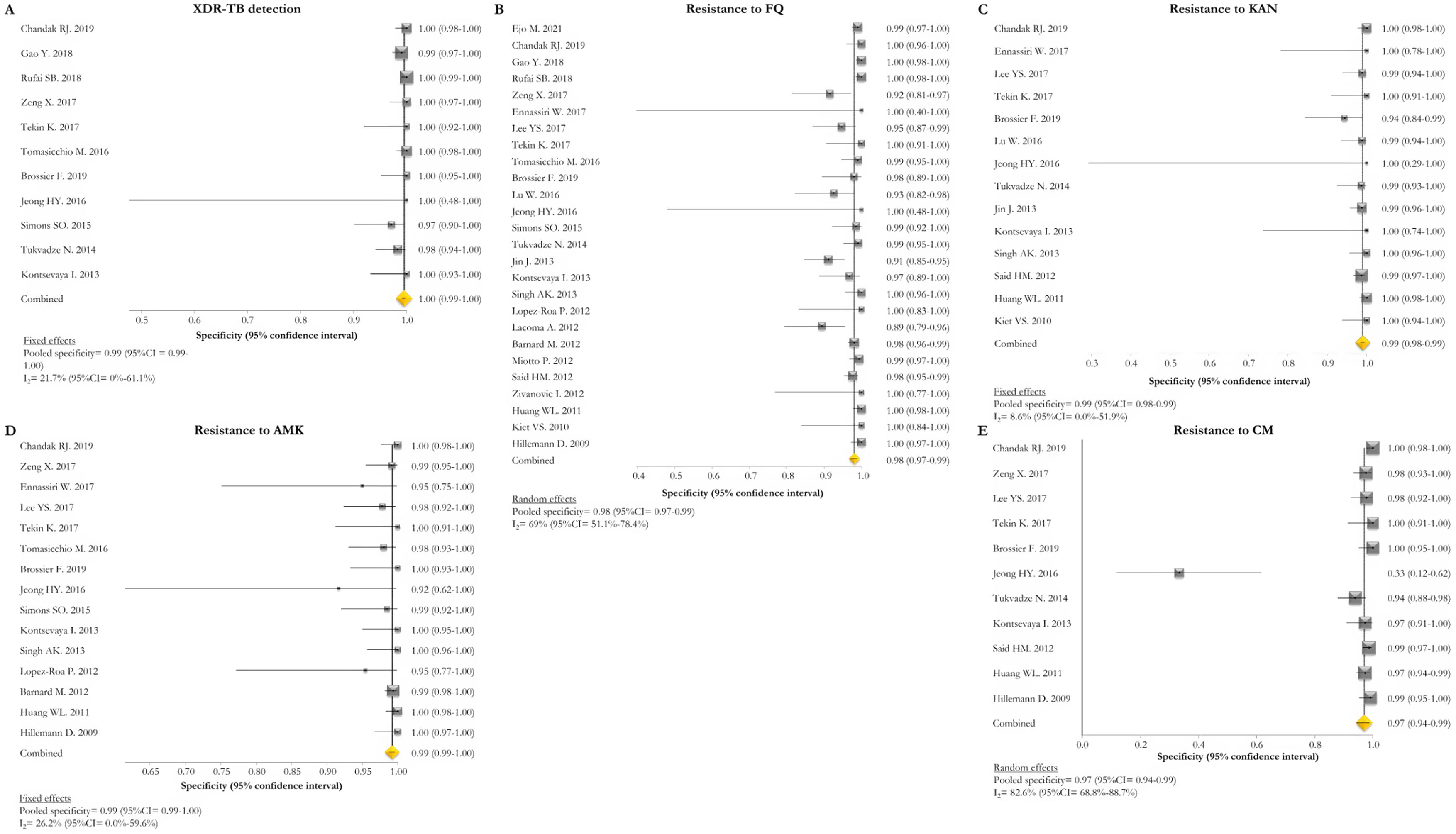
GenoType MTBDRsl VER 1.0The GenoType MTBDRsl VER 1.0 showed a pooled sensitivity and specificity for XDR-TB detection of 75% (95% CI=56–90; I2=91.5%) and 99% (95% CI=99–100; I2=21.7%), respectively, based on the evaluation of 11 studies.31–33,36,41,43,44,46,49,50,52 The pooled proportions of diagnostic sensitivity for resistance to FQ, KAN, AMK, and CM were 84% (95% CI=79–88; I2=80%), 64% (95% CI=46–80; I2=94%), 87% (95% CI=79–94; I2=79.9%), and 73% (95% CI=59–85; I2=85%), respectively. Furthermore, the pooled specificity for resistance to all tested drugs was higher than 95%: 98% (95% CI=97–99; I2=69%), 99% (95% CI=98–99; I2=8.6%), 99% (95% CI=99–100; I2=26.2%), and 97% (95% CI=94–99; I2=82.6%) for resistance to FQ, KAN, AMK, and CM, respectively (Figs. 5 and 6). Additional diagnostic performance variables of GenoType MTBDRsl VER 1.0 were summarized in Table S2.
Forest plots of pooled sensitivity of XDR-TB detection (A), and for resistance to FQ (B), KAN (C), AMK (D), and CM (E) of GenoType MTBDRsl VER 1.0. The point estimates of sensitivity from each study are indicated as a square and a 95% confidence interval is shown with a horizontal line; the yellow diamond is representative for the combined sensitivity.
Forest plots of pooled specificity of XDR-TB detection (A), and for resistance to FQ (B), KAN (C), AMK (D), and CM (E) of GenoType MTBDRsl VER 1.0. The point estimates of specificity from each study are indicated as a square and a 95% confidence interval is shown with a horizontal line; the yellow diamond is representative for the combined specificity.
The quality of included studies was assessed using the QUADAS-2 tool.24 Within the patient selection domain, 7 (18.4%) and 8 (21.1%) studies were evaluated as at high and unclear risk of bias, respectively. A high risk of bias was found in 4 studies in relation to the index and reference standard test domains (blinding evaluation). Moreover, 65.8% of the selected studies did not clarify if index test results were interpreted without previous knowledge of the reference standard results. All studies included a reference standard and were judged to have a low risk of bias in the flow and timing domains. No concerns emerged on the applicability of the study domains (Fig. S1).
DiscussionThe aim of our study was to assess all the tools able to rapidly diagnose pre-XDR/XDR-TB (and describe their technical characteristics), as well as to compare the diagnostic techniques and approaches in terms of sensitivity, specificity, laboratory complexity, time to confirmed diagnosis (Table 3).
Summary of the main indicators of diagnostic accuracy of the selected molecular techniques.
| Xpert MTB/XDR | GenoType MTBDRsl VER 2.0 | GenoType MTBDRsl VER 1.0 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| XDR | FQ | KAN | AMK | CM | XDR | FQ | KAN | AMK | CM | XDR | FQ | KAN | AMK | CM | |
| Sensitivity (%) | – | 94.0 91.4 | 86.0 98.1 | 73.0 91.0 | 61.0 70.0 | 85.0 | 95.0 | 89.0 | 90.0 | 89.0 | 75.0 | 84.0 | 64.0 | 87.0 | 73.0 |
| Specificity (%) | – | 99.0 98.5 | 98.0 97.4 | 100.0 98.9 | 100.0 99.7 | 98.0 | 98.0 | 98.0 | 100.0 | 99.0 | 99.0 | 98.0 | 99.0 | 99.0 | 97.0 |
The results of our study demonstrated that two rapid WHO-endorsed tests are available for these purposes, namely the Xpert MTB/XDR and GenoType MTBDRsl (VER1.0 and VER 2.0).
Xpert MTB/XDR test is a rapid, automated NAAT test at low complexity.21,22 It does not require any sophisticated laboratory infrastructure. The molecular amplification of small quantity of genetic materials takes place inside the cartridge and require less than 2h to provide results. The main advantages over the LPAs are the higher sensitivity for detecting Mycobacterium tuberculosis (it is also recommended in persons with a sputum smear-negative specimen), faster time to response (90min vs. 48–72h), simple handling, reduced number of invalid results for direct testing, its potential implementation outside a sophisticated laboratory, and its safety, as the cartridge represents ‘the laboratory’ where molecular amplification takes place. The Xpert MTB/XDR test can be done taking advantage of existing GeneXpert platforms equipped with 10-colour modules for Xpert MTB/RIF Ultra testing, or through system upgrade from 6-colour modules, with technical capacity to run the Xpert technology available yet on very large scale as infrastructure and procedures are equal. As technology upgrade may be logistically challenging and somehow expensive, different solutions have been conceived, including procurement of new 10-colour GeneXpert modules, new full systems, satellites (instrument only) to connect to existing GeneXpert system or converting an existing system with 6 colours modules to 10 colour modules. In terms of performance of the tests, they are very similar: in our study, it seems to slightly favour Xpert, although only 2 studies were available (sensitivity 91.4–94; specificity 98.5–99; accuracy 97.2–97.7; PPV 88.9–99.1; NPV 95.8–98.9).
The Xpert MTB/XDR is intended as a follow-on test after Xpert MTB/RIF or Xpert MTB/RIF Ultra testing, using left over sample reagent treated specimen, thus not necessarily requesting a second sample or culture to be collected. This approach is attractive with benefits for access to rapid DST, diagnostic laboratory workflow and turnaround time, or after other WHO-endorsed bacteriological tests confirming the presence of pulmonary TB. However, the performance of any rapid assay used as reflex test for a specimen previously determined to be M. tuberculosis positive and harbouring other resistance (e.g., rifampicin) will need to be carefully assessed in future studies, for if the initial rapid test is more sensitive than its reflex assay it may lead to invalid or non-actionable results and challenges with final interpretation of laboratory results.
A second set of considerations relates to the capacity of these tests to reliably diagnose XDR-TB, as newly defined by WHO in 2021.11 Xpert MTB/XDR detects M. tuberculosis complex DNA and mutations associated with resistance to isoniazid, fluoroquinolones (ofloxacin, moxifloxacin, levofloxacin, gatifloxacin), second-line injectable drugs (amikacin, kanamycin, capreomycin), and ethionamide in a single test, whereas GenoType MTBDRplus detects resistance to isoniazid and rifampicin, and GenoType MTBDRsl to fluoroquinolones (ofloxacin and moxifloxacin) and aminoglycosides/cyclic peptides (injectable antibiotics such as kanamycin, amikacin, capreomycin, and viomycin) in its 2 versions. Unfortunately, these tests are testing the drugs defining XDR-TB according to the previous 2006 WHO definition,64 which now reflects pre-XDR due to their capacity to detect resistance to fluoroquinolones, and not XDR-TB as neither these tests nor other genotypic DST assays can rapidly diagnose resistance to linezolid and bedaquiline. This is an important gap that will become more important with time; currently bedaquiline resistance is being detected in over 40% of cases failing the all oral regimen in South Africa, prevalence of bedaquiline resistance is currently 4% in South Africa and 15% in Moldova.65,66 Higher MICs to pretomanid have been detected in Lineage 1 strains.67 A 3 drug BPaL (bedaquiline, pretomanid, linezolid) regimen for pre-XDR with no rapid resistance test beforehand may therefore not be without risk and a shorter duration of lower dose linezolid may select for resistance to linezolid and thwart future salvage regimens. Targeted Next Generation Sequencing solutions are available but there is insufficient understanding of the role of resistance-conferring mutations to these drugs and such technologies are expensive and not implemented yet in many settings.68,69 Nonetheless, the two rapid assays provide valuable information at least on resistance to isoniazid and fluoroquinolones with now also the Xpert MTB/XDR technology and not only LPAs enabling to distinguish low- versus high-level resistance to these drugs.28 The detection of specific mutations is not implemented in the GeneXpert software automated interpretation. This information is nevertheless very relevant for clinicians who can rapidly design the best regimen with high precision guided by the knowledge of the association between specific genetic mutations and phenotypic resistance levels.70 For example, in the case where gyrA A90V, S91P, and D94A mutations associated with low-level fluoroquinolone resistance are identified by specific melting temperature window patterns, high dose moxifloxacin could still work. The detection of resistance to ethionamide and second-line injectables seems currently less clinically relevant, although these medicines can still be added to complete MDR regimens when other options are not available.
Considering the TB diagnostic algorithms and diagnostic characteristics, Xpert MTB/XDR could be placed at or near point-of-care settings to be used primarily as a follow-on test for laboratory-confirmed TB, complementing existing rapid tests that detect only rifampicin, or rifampicin/isoniazid resistance. Detection of fluoroquinolone resistance is essential to inform the use of all-oral 6–12 month standardized shorter regimen for MDR/rifampicin-resistant TB. The assay is conditionally recommended for the initial detection of resistance to isoniazid and fluoroquinolones, rather than culture-based phenotypic DST in people with bacteriologically confirmed pulmonary TB, and for the initial detection of resistance to these drugs and ethionamide and amikacin in people with bacteriologically confirmed pulmonary TB and resistance to rifampicin.17 Xpert MTB/RIF or Xpert Ultra and Xpert MTB/XDR could also be used as the initial diagnostic tests to detect TB, rifampicin, isoniazid and fluoroquinolone resistance in order to achieve universal DST; this algorithm may be preferable in settings with a high MDR-TB burden and also in those with high risk of isoniazid resistance (algorithm 117). If not feasible, it may be used for further evaluation of patients with rifampicin-resistant or MDR-TB, for patients with rifampicin-susceptible TB but no results available for isoniazid but at a high risk for isoniazid resistance (algorithms 3–4).
Our review is limited by high heterogeneity of the population targets and limited range of laboratory tools available. We also note limitations in the study designs of published work identified, including little clinical outcome data following treatment based on test findings.
Finally, rapid diagnostic tests are not only helpful in existing algorithms for guiding drug selection but may also guide dose selection (higher dose) in case of low-level resistance when limited alternative treatment options exist.20 Further testing options when a patient is demonstrating slower response to TB treatment than expected, despite proven drug susceptibility would be valuable, such as supplementary rapid tests for drug exposure. We foresee that the introduction of saliva- and urine-based point-of-care testing as well as microsampling techniques could be provided in conjunction with rapid DST as they are becoming more affordable for resource-limited settings, and may support laboratory testing efforts towards better outcomes for people affected by TB around the world.
FundingThe systematic review was partially funded via an unrestricted grant by Cepheid Europe SAS to the Public Health Consulting Group, Lugano, Switzerland. The donor had no role in conducting the systematic review, as well as analysing and interpreting the results and writing the manuscript.
Conflict of interestThe authors declare to have no conflict of interest directly or indirectly related to the manuscript contents.
The study is part of the scientific activities of the GTN (Global Tuberculosis Network).