CURRENT SITUATION AND LONG-TERM CONSEQUENCES OF COVID-19 INFECTION
More infoAll living organisms, in particular RNA viruses, are prone to mutation. It is the key of their evolution. During the COVID-19 pandemic, the worldwide capacity for sequencing SARS-CoV-2 in microbiology and public health laboratories has yielded enormous genetic information regarding the real time adaptation of this virus, and this race will continue as long as the virus infects new individuals. Many variants were detected from the first days in January to the end of February 2020. However, only a limited number of variants have had biological significance from the transmissibility, severity, or vaccine/immune escape point of view. These variants have been named from a public health perspective as “variants of concern” (VOC). With 2 years of perspective, we can associate each pandemic wave to the selection and dispersion of a new VOC (although this acronym was only included in Public Health reports from December 2020).
In February/March 2020, the SARS-CoV-2 variant responsible for the first pandemic (B.1) wave had two changes with respect to the wild-type “Wuhan-variant”, characterized by a mutation in the spike protein (D614G).1 This mutation appeared to enhance viral replication.2 Since then, practically all SARS-CoV-2 variant, carry this change. However, when SARS-CoV-2 was globally distributed, the selection of variants and pandemic waves occurred asynchronously in different regions. Obviously, the most notable variants were Alpha (detected in September 2020 in UK), Delta (October 2020 in India) and Omicron (November 2021 in South Africa and Botswana) associated in Spain with the third, fifth, and sixth pandemic waves, respectively.
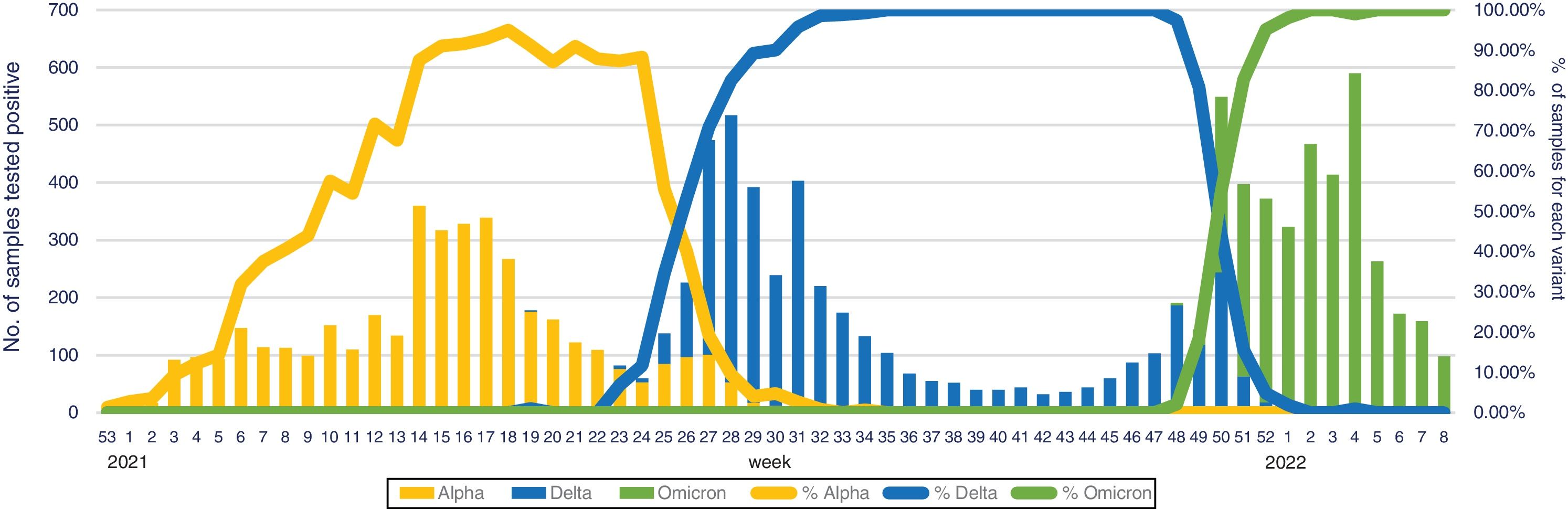
All these new VOC have a common characteristic, they are more transmissible than their parents, in some cases associated with a more potential virulent trait (Beta and Gamma variants), but on other occasions with less virulence (Omicron variant). Among them, the Omicron variant (B.1.1.529), with a doubling time of 2–3 days spreading speed,3 is much faster than the Delta variant (2×) and the original B.1 variant (10×).4 In South Africa, where Delta and Omicron variants co-existed during several weeks, the weekly increase of Omicron over Delta was 5.4-fold (95% confidence interval [95%CI], 3.1–10.1).3 This data revealed a short time between being infected and becoming infectious to other people (generation time [GT]) compared to other variants (GTAlpha, 5.5 days; GTDelta, 4.7 days, and GTOmicron, 3.2 days) or other highly transmissible viruses (GTmeasles, 12 days), and suggests that variant could be “the fastest-spreading virus in history” (title found in several general newspapers).5 These fears were confirmed a few weeks later, when Omicron was responsible for national incidences not previously described and were resumed by the WHO Director General on February 1st 2022 at the media briefing on COVID-19: “Since Omicron was first identified just 10 weeks ago, almost 90 million cases have been reported to WHO, more than were reported in the whole of 2020.” In fact, in our experience during the emergence and follow-up of different VOCs, it took nearly 20 weeks for the Alpha variant to reach the maximum peak since its first detection in our institution. This period was 15 weeks for the Delta variant and just 6 weeks for the Omicron (Fig. 1). Currently, in Europe, the Omicron variant represents >98% of sequenced samples and the Delta variant is still detected in only two countries: Latvia and Slovakia (https://www.ecdc.europa.eu/en/covid-19/country-overviews, last access 25 February 2022, corresponding to 6 weeks of 2022).
The observed increase in the infectivity of the Omicron variant is probably due to the high number of mutations (10–15 changes) found in the receptor-binding domain (RBD) compared to one or two in Alpha or Delta variants. Among these mutations, N501Y, T478K, and N440K mutations have a main role.6 N501Y is present in the Alpha variant and T478K is in the Delta variant. Despite the higher infectivity of Omicron, the hospitalization rate was 20–25% lower than the Delta variant.7 This could be related to its higher replicative capacity in the human bronchus, but 10 times lower in the human lung tissue,8 which may explain why it spreads so rapidly in human populations. This characteristic was previously observed in the Alpha variant, with which Omicron shares three spike-mutations (N501Y, P681H, and D614G), justifying the fast spread in the human population.9
On the other hand, the ability of Omicron to evade the COVID-19 vaccine immunity and cause breakthrough infections was also predicted to be higher than with previous variants, probably due to the presence of K417N, E484A, and Y505H mutations, the two first mutations being shared with the Beta variant (also described in South Africa in September 2020).10 Preliminary analyses suggest that, although the spike mutations involve several T cell and B cell epitopes of the immunological response, the majority of epitopes (>70%) remain unaffected, but initial laboratory data suggested that the Omicron variant is likely to weaken the COVID-19 vaccine protection. This observation could explain the high re-infection rate observed in vaccinated and previously infected people. Altarawnwh et al.11 found that the effectiveness of previous infection in preventing reinfection was estimated to be 90.2% against the Alpha variant, 85.7% against the Beta variant, 92.0% against the Delta variant, but only 56.0% against the Omicron variant, although the patients had high antibodies titres.12 Therefore, the Omicron variant is particularly insensitive to antibodies elicited against prior variants. Antigenic analyses indicated that all previous VOCs belong to one large antigenic cluster, whereas Omicron forms a new cluster, escaping vaccine or convalescent sera. For this reason, several authors have suggested considering Omicron as a new serotype.13 This proposal could have important implications for the development of updated vaccines, and also could improve our understanding of the cellular and humoral response.
Until today, several Omicron sub-variants have been identified. These include BA.1 (B.1.1.529.1), BA.2 (B.1.1.529.2), and BA.3 (B.1.1.529.3), which are all being monitored by WHO under the umbrella of ‘Omicron’. They share 31 mutations, and each one of them has 1–13 specific mutations. B.1 is the most widely distributed sub-variant, and is responsible for 98% of cases. BA.2 (1%) is increasing its weekly proportion. In several regions it has reached 50%, and in Denmark, 80%. Fortunately, the global circulation of all different variants is reportedly declining. BA.2, also called “stealth Omicron,” is 1.5 times as infectious as BA.1 and about 4.2 times as contagious as the Delta variant. A worrying data is that BA.2 has a 30% higher potential than BA.1 to escape existing vaccines.14 In fact, Israel reported a handful of cases of patients who were infected with the original Omicron BA.1 strain and have been reinfected with BA.2 in a short period. The description of these cases suggests that the antibodies generated from the early Omicron BA.1 were evaded by the BA.2 strain. To finish the bad news regarding BA.2, while monoclonal antibodies such as sotrovimab and tixagevimab retain significant neutralizing activity against BA.1, these may be not efficient against the BA.2. subvariant.
The comments of this editorial reveal that SARS-CoV-2 is continuously evolving in an endless race with the host, with new threats waiting a better epidemiological scenario. For this reason, we should not stop our efforts in SARS-CoV-2 surveillance, nor should we be surprised by the capacity of the viral evolution. We would like to finish with a new sentence of Tedros Ghebreyesus, WHO Director General: “It is premature for any country either to surrender or to declare victory.”
FundingJG research is supported by Instituto de Salud Carlos III and Ministerio de Ciencia e Innovación through CIBER en Salud Pública (CIBERES, CB06/02/0053) and RC through REIPI (RD16/0016/0011) and CIBER de Enfermedades Infecciosas, CIBERINFEC (CB21/13/00084).
Conflicts of interestNone.