Small cell lung cancer (SCLC) comprises 10–15% of all lung cancer cases and is the most aggressive histological type. Survival is poor and the molecular landscape of this disease is extraordinarily complex. The objective of this paper was to perform a Genome-Wide Association Study (GWAS) of this disease using a case–control study specifically designed for small cell lung cancer (SCLC).
MethodsIncident cases were consecutively recruited from 8 hospitals from different regions of Spain. Controls were recruited from the same hospitals using a frequency sampling based on age and sex distribution of cases. Biological samples were obtained along with detailed information on cases and controls lifestyle, including tobacco and radon exposure.
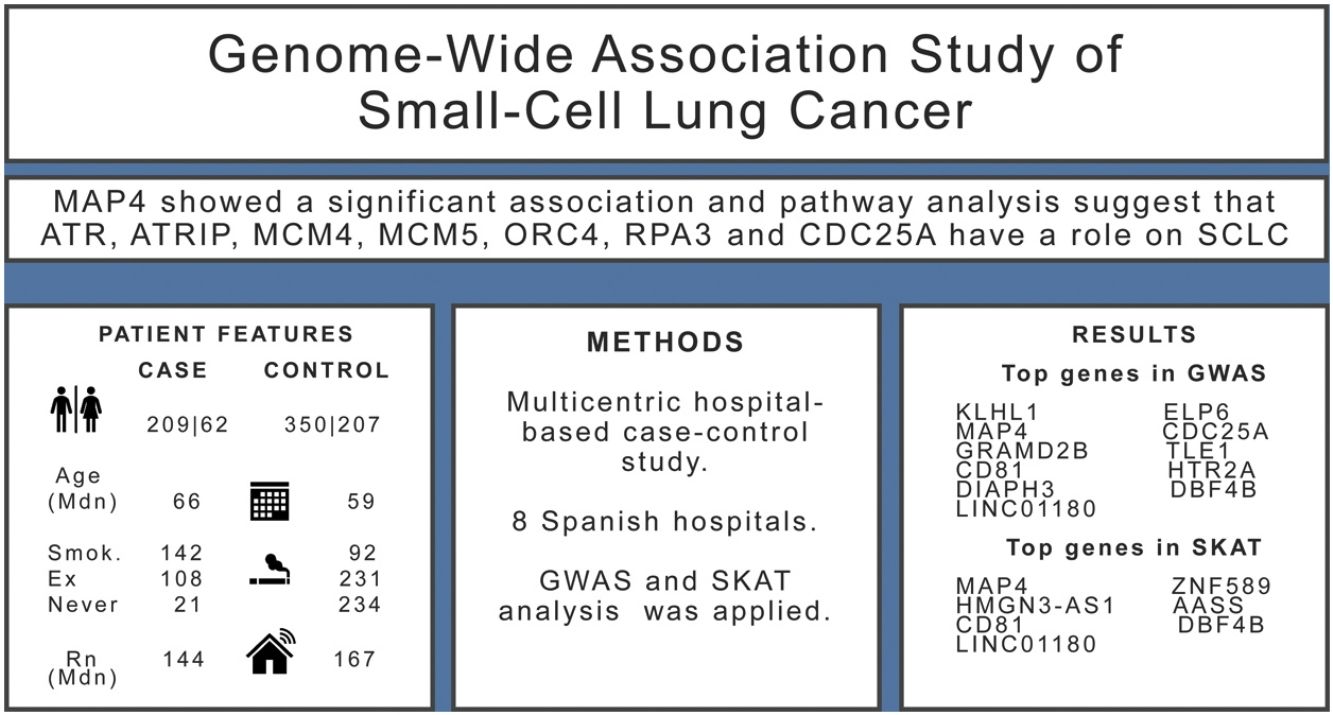
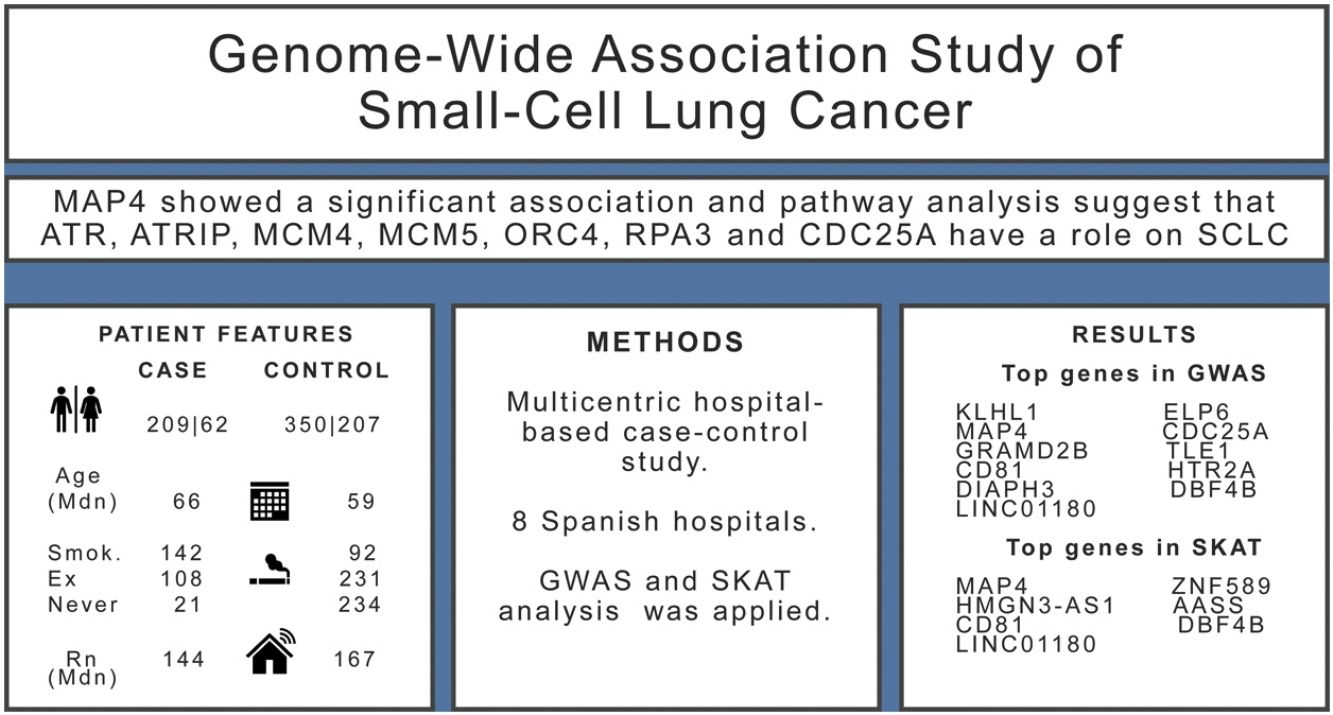
ResultsWe included 271 SCLC cases and 557 controls. We found evidence (p-values<10−5) of an association in the complete dataset for several loci, while MAP4 showed a significant association in the gene-based analysis. Pathway analysis suggested that ATR, ATRIP, MCM4, MCM5, ORC4, RPA3 and CDC25A genes have a role on the onset of SCLC.
ConclusionThis study provides biological evidence for pathways related to SCLC, offering novel loci for further research.
Lung cancer is the deadliest cancer, according to the International Agency for Research on Cancer (IARC). It comprises approximately 11.4% of all cancers and 18.4% of all cancer deaths.1 Despite recent advances including immunotherapy and targeted treatments directed to driver genes, 5-year survival remains low. In the USA, 5-year lung cancer survival is 22.9% for cancers diagnosed between 2012 and 2018.2
Small cell lung cancer (SCLC) is currently the most aggressive histological type and is considered to be a different entity when compared to other histological types. Approximately 10–15% of all lung cancers are diagnosed as small cell.2 The treatment of SCLC has seen few recent advances; successful treatment is infrequent with an estimated 5-year survival of 7%.3 SCLC is also the histological subtype most tightly linked to tobacco consumption.4 In addition, residential radon exposure has been associated with the occurrence of SCLC.5–7 Indoor radon has been classified by WHO and USEPA8 as the most important risk factor for lung cancer following tobacco consumption and the second most important risk factor in ever-smokers.
The molecular characteristics of SCLC are complex; general features include numerous chromosomal rearrangements and a very high mutational burden. Furthermore, inactivation of the tumor protein p53(TP53) and retinoblastoma transcriptional corepressor 1 (RB1) pathways is very common in SCLC.9 However, the persistent lack of specific and effective treatments for this disease paints a disheartening clinical picture and SCLC is still mainly treated with platinum-based chemotherapy.
Genome-Wide Association Studies (GWAS) have been found to provide important clues to improve our understanding of the role of genetic traits in causation (and thereby risk) of numerous diseases, and they have proved useful for identifying new genes and pathways associated with tumors. Identification of these novel pathways can also assist in the later development of new specific treatments. SCLC has been infrequently investigated using the GWAS approach as research efforts have been focused primarily upon the discovery of genetic loci linked to improve survival.10 To our knowledge, no GWAS study has been performed exclusively on SCLC and no study has adjusted its results for both indoor radon exposure as well as smoking status.
Here, we aimed to identify genetic loci associated with SCLC using a GWAS approach applied in a case–control study performed in Galicia, a radon-prone area located in North-western Spain.
MethodsDesign and settingWe conducted a multicentric, hospital-based, case–control study in several Spanish regions with 8 hospitals recruiting patients. The recruitment took place between September 2015 and August 2019. All cases were incident, with a pathologically confirmed diagnoses of small cell lung cancer and were recruited consecutively. Controls were collected from the patient population of the same recruiting hospitals for minor surgery not related to tobacco consumption (i.e. inguinal hernias, other minor surgeries, etc.). To be included, both cases and controls had to be older than 30 without an upper age limit. Controls were recruited using sex and age frequency matching to cases to assure a similar distribution of both variables. The study protocol was approved by the Santiago de Compostela-Lugo Ethics Committee (REF 2015/222). We followed the STROBE guidelines for the communication of results of observational studies.11
Information retrieval and radon measurementsAll participants were personally interviewed by trained research staff members. The interview was focused on specific lifestyle habits with special emphases on tobacco consumption. Occupational history also was obtained. A radon detector was given to all participants to be placed in their home for at least 3 months. Instructions for placement of detectors were provided and all participants were phoned twice, the first time to assure that the detector was correctly placed and the second to remind participants to send back the detector, properly sealed, to the laboratory. Detectors were placed in the main bedroom following standard instructions. All devices were read at the Galician Radon Laboratory, located at the Hospital Complex of Santiago de Compostela.
Biological samples and genotypingAll participants provided 3ml of peripheral blood which was frozen at −80°C until analysis. DNA samples were genotyped at the Spanish National Genotyping Center (CeGen-USC; Santiago de Compostela, Spain), using the Axiom Precision Medicine Research array, following the manufacturer's instructions (Axiom™ 2.0 Assay 96-Array Format Manual Workflow; ThermoFisher Scientific). Briefly, total genomic DNA (200ng) was amplified and randomly fragmented into 25–125 base pair (bp) fragments, which were then purified and re-suspended in a hybridization cocktail. The hyb-ready targets were then transferred to the GeneTitan Multichannel Instrument for automated, hands-free processing (including hybridization to Axiom array plates, staining, washing and imaging). CEL files were automatically processed for allele calling and quality control with the Axiom GT1 algorithm available through the Axiom Analysis Suite v4.0.3.3 and following the Axiom™ Genotyping Solution Data Analysis User Guide (ThermoFisher Scientific).
Quality control and imputationA quality control (QC) procedure was conducted at both single nucleotide polymorphism (SNP) and individual level using PLINK 1.912 and a custom R script. Variants were excluded according to the following criteria: minor allele frequency (MAF)<1%, call rate<98%, a difference in missing rate between cases and controls>0.02, or a deviation from Hardy–Weinberg equilibrium (HWE) expectations (p<1×10−6 in controls, p<1×10−10 in cases. Samples were removed from the analysis when the call rate was <98% or when the heterozygosity rate deviated more than 5 standard deviations from the mean heterozygosity of all individuals.
Principal components were calculated to control for population structure and identity by descent was estimated to assess kinship. Samples deviating more than 5 standard deviations from the mean value for the first two principal components were excluded. After quality control was completed, 361 cases and 743 controls remained, each with a total of 428,206 variants.
Imputation was conducted based on the TOPMed version r2 reference panel (GRCh38)13 in the TOPMed Imputation Server. After post-imputation filtering (Rsq>0.3, HWE p>1×10−6, MAF>0.01) 9,271,137 markers remained.
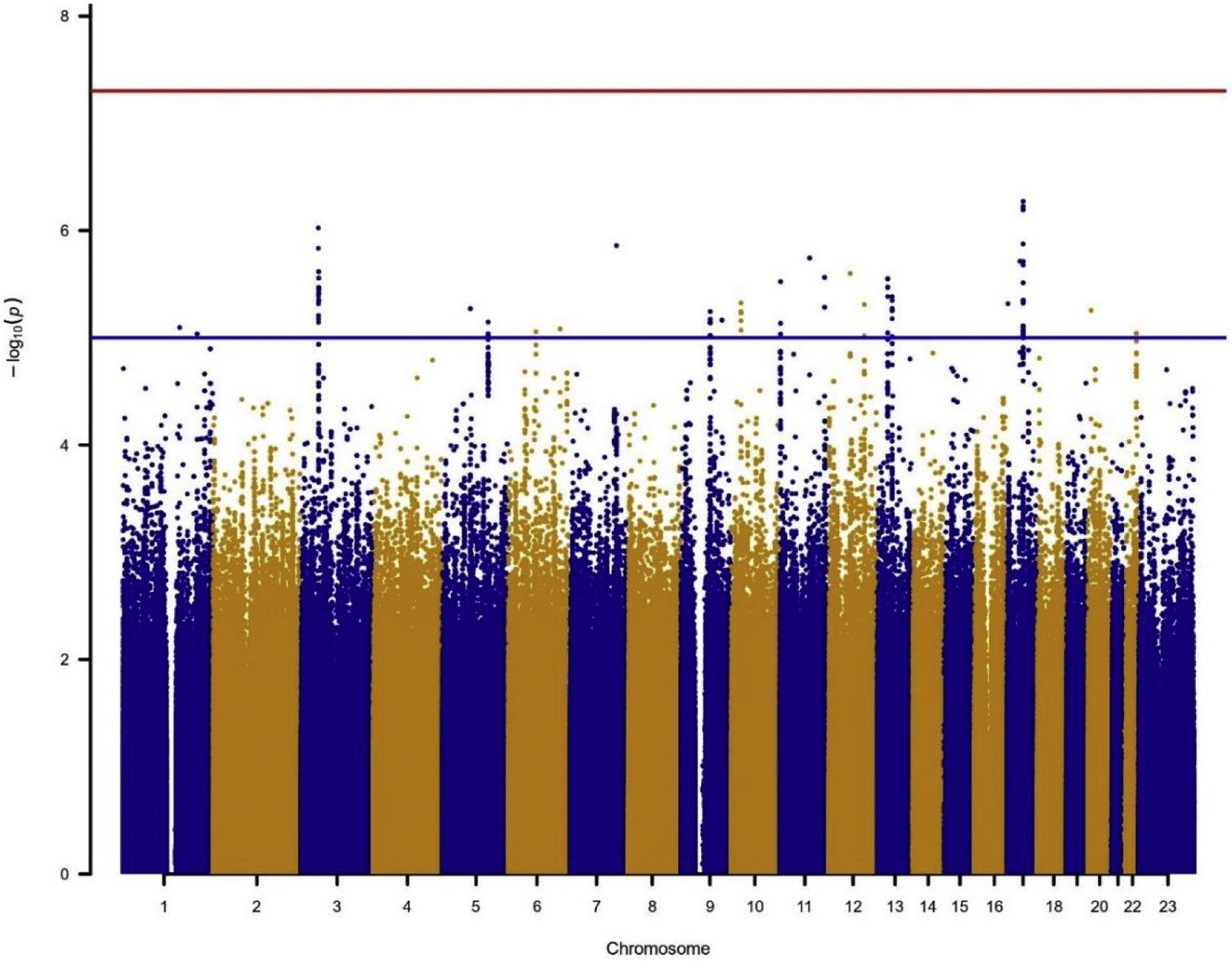
Association testingTo assess the genetic association, logistic mixed regression models were fit for each SNP (MAF>1%) under the additive model (genotypes coded as 0, 1 or 2 in function of the number of minor alleles) and using a case/control status as the dependent variable. All models were adjusted by age, sex, radon exposure, education level, tobacco consumption and the first 3 genetic principal components as covariates. Analyses were conducted in the complete dataset (Ncontrols=557, Ncases=271) and in the subgroup of smokers (Ncontrols=323, Ncases=250). Analyses in never-smokers were not feasible due to the low number of cases. R libraries SNPRelate14 and SAIGEgds15 were used for this purpose. Significance was established at p<5×10−8, and Manhattan plots showing the −log10 (p-value) for each SNP were obtained with the R library qqman.16 Representative SNPs of the regions showing suggestive evidence of association (p-values<10−5 for more than 2 markers) were selected using the clump function of PLINK 1.9 (clumping parameters r2=0.5, p1=5×10−8 and p2=0.05).
Complementarily to the GWAS analysis, the Sequence Kernel Association Test (SKAT)17 was applied to assess the association between the phenotype and the combined effect of variants in each gene. Four different approaches were used: (i) combining the effect of rare and common variants using equal weights for all variants (SKAT_w1), (ii) using default SKAT weights (higher weight to rarer variants) for all variants (SKAT), (iii) combining only low frequency variants (MAF<0.05, SKAT_low), and (iv) using a simpler collapsing method for all variants, the burden test (BURDEN). Markers that remained after the post-imputation QC were aggregated into gene sets, and only those sets comprising at least 3 markers were analyzed, resulting in a total of 21,732 genes. The Bonferroni correction was set at p<2.3×10−06 for α=0.05. SKAT tests were adjusted for the covariables age, sex, radon exposure, education level and the first 3 principal components.
Finally, a pathway analysis was performed for the list of genes showing a SKAT p<0.01, using WEB-based Gene SeT AnaLysis Toolkit.18
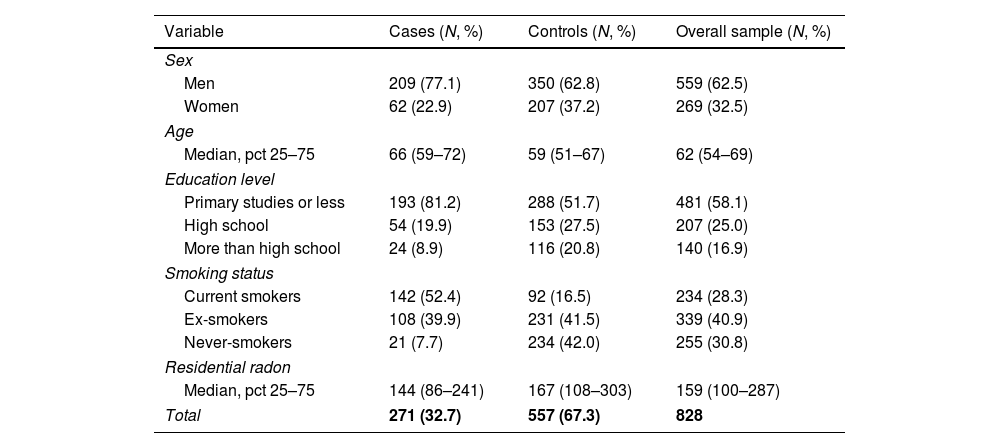
ResultsThe sample size available for the GWAS analyses included 271 cases and 557 controls. Among cases, 77% were men compared with 63% in controls, and cases were slightly older than controls (Table 1). Tobacco consumption was more frequent and more intense in cases compared to controls. Indoor radon concentrations were slightly lower in cases compared to controls. A sample description by case and control status can be found in Table 1.
Patient characteristics by case and control status.
| Variable | Cases (N, %) | Controls (N, %) | Overall sample (N, %) |
|---|---|---|---|
| Sex | |||
| Men | 209 (77.1) | 350 (62.8) | 559 (62.5) |
| Women | 62 (22.9) | 207 (37.2) | 269 (32.5) |
| Age | |||
| Median, pct 25–75 | 66 (59–72) | 59 (51–67) | 62 (54–69) |
| Education level | |||
| Primary studies or less | 193 (81.2) | 288 (51.7) | 481 (58.1) |
| High school | 54 (19.9) | 153 (27.5) | 207 (25.0) |
| More than high school | 24 (8.9) | 116 (20.8) | 140 (16.9) |
| Smoking status | |||
| Current smokers | 142 (52.4) | 92 (16.5) | 234 (28.3) |
| Ex-smokers | 108 (39.9) | 231 (41.5) | 339 (40.9) |
| Never-smokers | 21 (7.7) | 234 (42.0) | 255 (30.8) |
| Residential radon | |||
| Median, pct 25–75 | 144 (86–241) | 167 (108–303) | 159 (100–287) |
| Total | 271 (32.7) | 557 (67.3) | 828 |
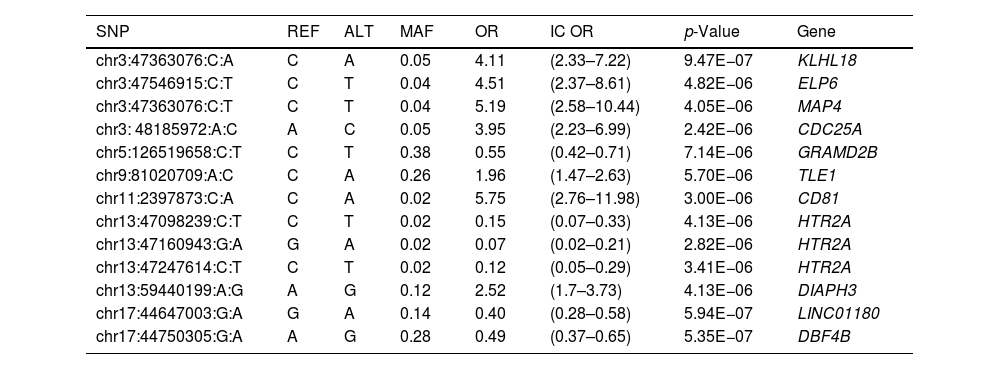
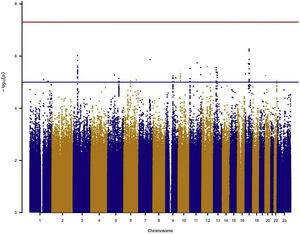
In the individual association analysis performed on the complete dataset, no SNP reached genome-wide significance. However, several borderline significant loci (p-values<10−5) were observed in the Manhattan plots (Fig. 1). Table 2 shows the top independent SNPs with suggestive evidence of an association in the complete dataset. Interestingly, the results for the GWAS of ever-smokers revealed the same loci as the GWAS with all the individuals (shown in Supplementary Table 1 and Supplementary Fig. 1).
Manhattan plot for the GWAS analysis including smokers and non-smokers, adjusting for sex, age, 3 PCs, tobacco consumption, education level and radon exposure. Manhattan plot represents, in chromosomic order (x-axis), the p-values of the entire GWAS on a genomic scale and the −log10 of this value (y-axis). The genome-wide significance level is plotted as the red line and the lower limit of the “shadow region” is plotted as the blue line.
Top independent SNPs for the GWAS analysis in the general group, including both smokers and non-smokers (p-value<1×10e−5).
| SNP | REF | ALT | MAF | OR | IC OR | p-Value | Gene |
|---|---|---|---|---|---|---|---|
| chr3:47363076:C:A | C | A | 0.05 | 4.11 | (2.33–7.22) | 9.47E−07 | KLHL18 |
| chr3:47546915:C:T | C | T | 0.04 | 4.51 | (2.37–8.61) | 4.82E−06 | ELP6 |
| chr3:47363076:C:T | C | T | 0.04 | 5.19 | (2.58–10.44) | 4.05E−06 | MAP4 |
| chr3: 48185972:A:C | A | C | 0.05 | 3.95 | (2.23–6.99) | 2.42E−06 | CDC25A |
| chr5:126519658:C:T | C | T | 0.38 | 0.55 | (0.42–0.71) | 7.14E−06 | GRAMD2B |
| chr9:81020709:A:C | C | A | 0.26 | 1.96 | (1.47–2.63) | 5.70E−06 | TLE1 |
| chr11:2397873:C:A | C | A | 0.02 | 5.75 | (2.76–11.98) | 3.00E−06 | CD81 |
| chr13:47098239:C:T | C | T | 0.02 | 0.15 | (0.07–0.33) | 4.13E−06 | HTR2A |
| chr13:47160943:G:A | G | A | 0.02 | 0.07 | (0.02–0.21) | 2.82E−06 | HTR2A |
| chr13:47247614:C:T | C | T | 0.02 | 0.12 | (0.05–0.29) | 3.41E−06 | HTR2A |
| chr13:59440199:A:G | A | G | 0.12 | 2.52 | (1.7–3.73) | 4.13E−06 | DIAPH3 |
| chr17:44647003:G:A | G | A | 0.14 | 0.40 | (0.28–0.58) | 5.94E−07 | LINC01180 |
| chr17:44750305:G:A | A | G | 0.28 | 0.49 | (0.37–0.65) | 5.35E−07 | DBF4B |
SNP: single nucleotide polymorphism; REF: reference nucleotide; ALT: altered nucleotide; OR: odds ratio; IC OR: odds ratio confidence interval.
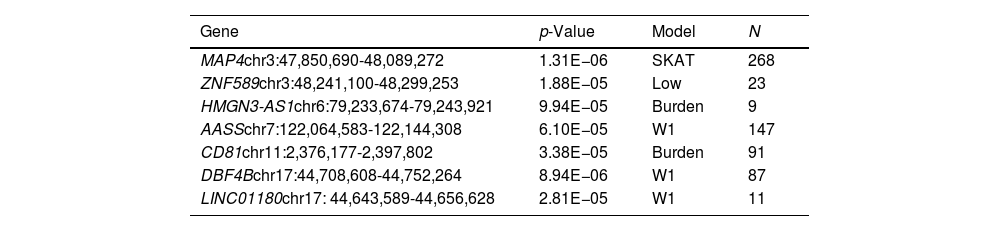
Results for the SKAT analysis are displayed in Table 3 and Supplementary Table 2 for the complete group and the ever-smokers’ group, respectively. Microtubule associated protein 4(MAP4) was associated with SCLC in this analysis (Table 3) and it is worth noting that this region was also borderline significant in the GWAS at SNP level (Table 2).
Top genes for SKAT analyses in smokers and non-smokers (p-value<1×10e−5).
| Gene | p-Value | Model | N |
|---|---|---|---|
| MAP4chr3:47,850,690-48,089,272 | 1.31E−06 | SKAT | 268 |
| ZNF589chr3:48,241,100-48,299,253 | 1.88E−05 | Low | 23 |
| HMGN3-AS1chr6:79,233,674-79,243,921 | 9.94E−05 | Burden | 9 |
| AASSchr7:122,064,583-122,144,308 | 6.10E−05 | W1 | 147 |
| CD81chr11:2,376,177-2,397,802 | 3.38E−05 | Burden | 91 |
| DBF4Bchr17:44,708,608-44,752,264 | 8.94E−06 | W1 | 87 |
| LINC01180chr17: 44,643,589-44,656,628 | 2.81E−05 | W1 | 11 |
Model: approach model used complementary to the GWAS analysis; N: number of cases; MAP4: microtubule associated protein 4; ZNF589: zinc finger protein 589; HMGN3-AS1: HMGN3 antisense RNA 1; AASS: aminoadipate-semialdehyde synthase; CD81: CD81 molecule; DBF4B: DBF4 zinc finger B; LINC01180: long intergenic non-protein coding RNA 1180.
Other two top genes from the SKAT analysis, CD81 molecule (CD81) and DBF4 zinc finger B (DBF4B), were also borderline significant in the GWAS or are located in the same chromosome region as GWAS signals (such as the Long intergenic non-protein coding RNA 1180 (LINC01180) regarding DBF4B or Zinc finger protein 589 (ZNF589) regarding MAP4. Results for smokers were not significant but the top genes were shared with the complete group analysis (Supplementary Table 2).
Pathway analysis was performed with the list of 522 genes showing a p-value<0.01 for any SKAT model in the entire study group. Over-representation analysis showed a significant enrichment (7 out 37 genes) for the Reactome pathway Activation of ATR serine/threonine kinase (ATR) in response to replication stress (p=1.1×10−05, FDR q=0.0199). The top SKAT genes contributing to this pathway were ATR, ATR interacting protein (ATRIP), minichromosome maintenance complex component 4 (MCM4), minichromosome maintenance complex component 5 (MCM5), origin recognition complex subunit 4 (ORC4), replication protein A3 (RPA3) and cell division cycle 25A (CDC25A). When this analysis was performed in the ever-smokers’ group, 613 genes were selected (p<0.01 in SKAT) and the same pathway was enriched (8 out 37 genes, p=5×10−06, q=0.0087).
DiscussionHere, we have presented a GWAS analysis in SCLC patients including, for the first time, adjustment for indoor radon exposure. Some gene loci with a role in proliferation and cell-cycle functions were found, suggesting association with SCLC risk, and the same signals were found when smokers were analyzed separately. The results were internally consistent, especially for signals of chromosome 3 and 17, when gene-based analysis was performed, suggesting overall that these may be variant loci associated with small cell lung cancer onset.
The genetic landscape of SCLC is perhaps the most complex of all lung cancer histological types. Alterations in both the TP53 and RB1 gene are the most frequent molecular changes observed in this lung cancer subtype.19 It has been suggested that functional inactivation of both genes is a pre-requisite for SCLC.9,20 There are also reports of differences between different subpopulations, with some studies indicating that mutations found in SCLC differ between Europeans and East Asians.20
To date, there are surprisingly few studies performed on SCLC using a GWAS approach. Most of these studies have a case–control design performed on populations of Chinese descent. To our knowledge, there have been only 3 studies that included European participants exclusively,10,21,22 as did our study. These studies included approximately 2000 SCLC cases, but were not designed specifically to examine SCLC as they were pooling studies. Two of these studies reported an association of a variant in the cholinergic receptor nicotinic alpha 3 subunit (CHRNA3) gene with SCLC.10,22 This gene encodes a member of the nicotinic acetylcholine receptor family of proteins and, similar to the data from the study of non-small cell lung cancer (NSCLC), may be involved in the onset of the higher risk of SCLC posed by heavy smoking. It has been shown that nicotinic acetyl choline receptors may mediate lung cancer growth.23 These genes are necessary for the viability of SCLC and nicotine promotes SCLC cell viability. Nevertheless, our data did not find any association of SCLC with this gene. This may be due to sample size, chance or could be attributable to adjustment for smoking and radon exposure. At the same time, it is important to highlight the fact that the results of our GWAS analysis were similar for the both the entire sample and the sample restricted to ever-smokers.
A large region in chromosome 3 was associated with SCLC in both analyses. This region involved the genes kelch like family member 18 (KLHL18), protein tyrosine phosphatase non-receptor type 2 (PTPN23), elongator acetyltransferase complex subunit 6 (ELP6), MAP4 and CDC25A. Whilst no markers reached the significance threshold in the GWAS, MAP4 was significantly associated in the gene-based analysis (Table 3). MAP4 plays an important role in microtubule assembly and stabilization. Expression levels of MAP4 protein in lung adenocarcinoma tissues was found to be significantly higher than those in noncancerous tissues.24 MAP4 expression was significantly correlated with differentiation, pathological T stage, and TNM stage and survival. Mutation in this gene occurs in approximately 13% of SCLCs arising in Asians.20 It has also been associated with melanoma and breast cancer25 and has been recently related with prognosis of NSCLC patients receiving atezolizumab.26
This region on chromosome 3 is extremely interesting since KLHL18 has been recently related with poor progression in a study performed in NSCLC cells.27 The in vitro analysis of NSCLC cells showed that overexpressing KLHL18 inhibited cell proliferation, migration, and invasion and some interrelationship with the expression of programmed death ligand 1 (PD-L1) protein can be present preventing tumor cell immune escape and therefore might be also associated response to treatments targeting PD-L1.27CDC25A alteration is commonly observed in the pathogenesis of lung cancer. A recent study has shown that CDC25A could be used to distinguish normal from SCLC samples, as CDC25A was overexpressed in SCLC samples.28 Other studies have reported that this gene may be involved in different processes of cell division and be a target for future treatments for NSCLC.29CDC25A was also one of the contributing genes to the ATR activation pathway found significant in the enrichment analysis.
A second suggestive signal was found in chromosome 17, including the genes DBF4B, LINC01180 and ADAM metallopeptidase domain 11(ADAM11). The marker with the lowest association p-value in the complete group is located within this region, chr17:44750305:G:A (Table 2). To our knowledge, there is only one study having associated DBF4B gene with cancer, and in this case with colorectal cancer. The DBF4B gene is involved in the regulation of the S-phase of the cell-cycle and is related with Cdc7 kinase.30
The SNP with the lowest p-value in the GWAS with ever-smokers was chr13:47160943:G:A (p=1.6×10−07, Supplementary Table 1). This SNP was also found among the top SNPs of the complete group (p=2.82×10−06, Table 2). The corresponding gene is 5-hydroxytryptamine receptor 2A (HTR2A), which codifies for the serotonin receptor and has been associated with different pathways related to pain and with basal cell carcinoma after irradiation,31 but, to our knowledge, no study has directly associated this gene with lung cancer.
The results obtained in the GWAS analysis were consistent with the gene-based analyses. MAP4 was the only gene with a significant association. Some other top SKAT genes were also pointed as suggestive signals in the GWAS (CD81 in the chromosome 11, DBF4B and LINC01180 in the chromosome 17, see Table 2) or located close to genes discussed in the GWAS, such as ZNF589 in relation to MAP4.
ZNF589 has been associated together with other genes (in a gene-clustering mode) with a worse progression of squamous cell lung carcinoma.32ZNF589 has been associated with several fundamental processes such as cell proliferation, apoptosis, differentiation, and tumorigenesis. This gene has also been associated with poor survival in breast cancer33 and with progression of digestive system carcinoma.34
Finally, the ATR activation pathway was found to be significantly enriched, and this pathway has been linked to cancer proliferation. ATR encodes RAD3 and ATM-related kinase involved in the cell-cycle that activates in response to DNA damage and to replicative stress, arresting the cell-cycle to avoid apoptosis. The role of CDC25A in this pathway is well known: through ATR-mediated CHK1 activation, the degradation of CDC25A is induced and thus the cell does not entry the replication phase.35ATR signaling has been established as a potential target for cancer therapy, as it promotes survival of tumor cells.36 In fact, ATR inhibition (in combination with DNA topoisomerase I (TOP1) inhibition) might be a therapeutic strategy for enhancing antitumor immunity in a (STING-related) subtype of SCLC.37
The present study has a series of advantages. Perhaps the main one is that, to our knowledge, it is the first GWAS performed exclusively for SCLC. A further advantage is that we have detailed information on tobacco consumption, something not always available, particularly in pooling studies. Further, a significant strength of the study is that we have collected and adjusted the results for measured residential radon exposure. Lastly, having recruited incident SCLC cases from different hospitals from distinct Spanish regions provides higher external validity.
This study also has several limitations. The main one is associated with the small sample size, although the lack of statistical power to detect significant associations for low frequency variants is minimized by performing gene-level analyses. Nevertheless, these results can provide some indication that several novel genes, which we identified, might be involved in small cell lung cancer onset. Most of our cases and controls are ever-smokers, and therefore our conclusions may not be valid for never-smoking lung cancer cases. Finally, an analysis broken down by sex is not possible due to sample size limitations.
To conclude, we observed that several different gene loci may be involved in the occurrence of small cell lung cancer. A multigenic approach is clearly needed to provide more information on the molecular pathways involved on this cancer. Our work adds important data to the extremely sparse literature exclusively targeting this specific histologic type of lung cancer. Most of the loci we observed to be associated with SCLC are related with the cell-cycle, and some of them have been linked previously with lung cancer, but none with SCLC. These loci could be explicitly followed up in additional studies with sample size enough to obtain some robust results. Finally, further studies should adjust their results by tobacco and residential radon exposure, the two main risk factors of this aggressive lung tumor.
Authors’ contributionJR Enjo-Barreiro wrote the first draft of the manuscript. Alberto Ruano-Ravina and Mónica Pérez-Ríos had the idea of the hypothesis tested and designed this research. All other authors contributed equally to this manuscript, critically reviewed different versions, approved the final version and take public responsibility on its content.
FundingThis work was supported by PI15/01211 – ISCIII – co-financed FEDER.
This research is part of the PhD work of José Ramón Enjo-Barreiro.
Conflict of interestsKarl Kelsey is a founder and scientific advisor to Cellintec, which had no role in this research.